Single Nucleotide Polymorphisms of NLRP12 Gene and Association with Non-specific Digestive Disorder in Rabbit
Article information
Abstract
The NLRP12 (NLR family, pyrin domain containing 12) serves as a suppressor factor in the inflammatory response and protects the host against inflammation-induced damage. In the present study, we aimed to study the polymorphisms of NLRP12 gene and its association with susceptibility to non-specific digestive disorder (NSDD) in rabbits. We re-sequenced the entire coding region of the rabbit NLRP12 gene and detected a total of 19 SNPs containing 14 synonymous and five non-synonymous variations. Among them, the coding SNP (c.1682A>G), which would carry a potential functional implication, was subsequently subjected to genotyping for case-control association study (272 cases and 267 controls). The results revealed that allele A was significantly protective against NSDD with an odds ratio value of 0.884 (95% confidence interval, 0.788 to 0.993; p = 0.038). We also experimentally induced NSDD in growing rabbits by feeding a fibre-deficient diet and subsequently investigated NLRP12 mRNA expression. The mRNA expression of NLRP12 in healthy status was significantly higher than that in severe NSDD (p = 0.0016). The highest expression was observed in individuals carrying the protective genotype AA (p = 0.0108). These results suggested that NLRP12 was significantly associated with the NSDD in rabbits. However, the precise molecular mechanism of NLRP12 involving in the development of rabbit NSDD requires further research.
INTRODUCTION
Digestive disorder in farm animals is a huge hindrance to the development of animal husbandry. In China, large-scale rabbit production has developed quickly in recent years. However, rabbits are very susceptible to various infections and have high mortality, especially after weaning. Among them, the intestinal disease is one of most common diseases and results in considerable economic losses in industrial rabbit farms (Licois, 2004). On the whole, intestinal disease could be separated into the specific enteropathies and non-specific digestive disorder according to whether the specific pathogens are present (De Rochambeau et al., 2006; Dewrée et al., 2007). The enteropathies could be induced by specific bacteria, viruses, and intestinal parasites. However, it is difficult to find any specific pathogen in a non-specific digestive disorder. Both the environmental and genetic factors are believed to determine the occurrence and development of non-specific digestive disorder (Bennegadi et al., 2001; De Rochambeau et al., 2006). Consequently, it is urgent to explore the genetic markers associated with non-specific digestive disorder in farm animals.
In animals, the innate immunity system acts with considerable specificity and is proficient at discrimination between pathogens and self. Special molecular signatures belonging to pathogens have been proposed as pathogen-associated molecular patterns (PAMPs), which can be recognized by the innate immune system (Janeway Jr and Medzhitov, 2002). In the process of host immune evolution, the receptors that recognize these PAMPs are referred to as pathogen recognition receptors (PRRs). A subtle ‘microbial sensing’ system that mediates pathogen recognition, consists of the Toll-like receptor (TLRs) and the nucleotide-binding domain leucine-rich repeat containing (NLR) family. The NLR family also functions as important components of the inflammation and immunity system (Ye and Ting, 2008). Recently, an increasing number of papers concerning human inflammatory diseases have focused on NLRP12, a member of the NLR gene family.
NLRP12 acts as negative regulator of inflammation, and this function requires the assistance of ASC (apoptotic speck protein) (Wang et al., 2002). ASC is a protein comprised of an N-terminal Pyrin domain and a C-terminal CARD domain (Richards et al., 2001). NLRP12 puts a damper on both canonical and noncanonical NF-κB activation and subsequent inhibition of production of proinflammatory cytokines and chemokines (Williams et al., 2005; Lich et al., 2007).
NLRP12 is a checkpoint in the process of many inflammatory diseases. Previous studies identified NLRP12 mutations in patients with periodic fever syndromes (Jeru et al., 2008; Jéru et al., 2011). Another research demonstrated significant association between NLRP12 mutation and atopic dermatitis (AD) (Macaluso et al., 2007). In a mouse model, NLRP12-deficient mice are highly susceptible to colon inflammation and tumorigenesis (Md. Hasan Zaki et al., 2011). The latest research also indicated that NLRP12 suppresses colon inflammation in mice (Allen et al., 2012). Altogether, NLRP12 plays a critical role in sustaining internal environment homeostasis in humans and animals. However, there is no report concerning association between NLRP12 polymorphism and digestive disorder in farm animals. In this study, we intended to explore the association between the polymorphism of NLRP12 gene and non-specific digestive disorder (NSDD) in rabbits.
In contrast to Mendelian disease, complex traits are comprehensive consequence of a number of factors both genetic and environmental. To search the genome locus and gene causing complex traits has become the focus of genome-wide association studies. Significant differences in allele frequencies between cases and controls are demonstrated as evidence for participation of an allele in disease susceptibility (Tang et al., 2005). In this study, we performed the case-control study to explore the association between NLRP12 polymorphisms and NSDD in New Zealand White rabbits.
MATERIALS AND METHODS
Ethics statement
Animal care and tissue collection procedures involved in the present study were sought and approved by the Institutional Animal Care and Use Committee in College of Animal Science and Technology, Sichuan Agricultural University, Sichuan, China (DKY-B20090908).
Recruitment of case and control animals
The general commercial population of New Zealand White rabbits was reared at the experimental research farm of Sichuan Agricultural University. After weaning at 28 d of age, rabbits were fed pelleted food (16% protein, 10.8 MJ/kg) until 84 d of age. The food was restricted to approximately 80% of average ad libitum intake and water ad libitum.
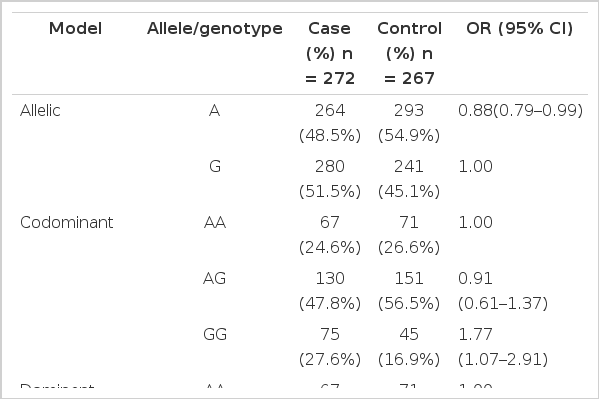
The recruitment process of case and control subjects has been addressed in detail in our previous report (Zhang et al., 2011). Briefly, a case group was composed of rabbits whose death were caused by NSDD from 28 to 84 d of age. According to clinical autopsy, only these individuals showing symptoms of watery content, dilatation, impaction, congestion and mucus in gastrointestinal tract were finally included in case group. In the same population, healthy rabbits without any clinical records of NSDD were included in control group by taking into account matching for sex. We also avoided a genetic relationship within three generations for all animals according to pedigree records. In total, 272 and 267 unrelated rabbits were finally collected in case and control groups, respectively (Table 4).
Experimentally fibre-deficient induced non-specific digestive disorder and collection of enteric tissues
Additional 60 healthy growing rabbits at ∼50 d of age, when the intestinal flora has fully established, were randomly selected. We experimentally induced the digestive disorder in these rabbits by feeding fibre-deficient diet, which is one of the most considerable risk factors (Bennegadi et al., 2001). NLRP12 mRNA expression and association with NSDD were subsequently investigated. After a 7-d dietary transition, these rabbits were fed fibre-deficient diet (CF = 9%, in contrast to the standard diet of CF = 15%) during the experimental period of two weeks.
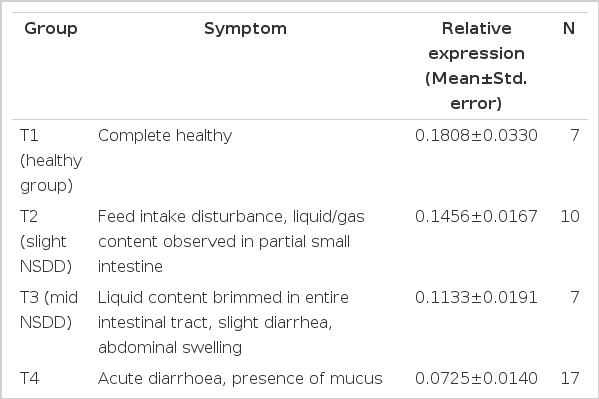
Animals were carefully observed twice a day with precise records kept for all clinical signs of digestive disorder, such as reduced feed intake, diarrhea, constipation (caecal impaction), and presence of mucus in excreta. At the end of experimental period, rabbits were slaughtered by intravenous administration of sodium pentobarbital and subsequently subjected to an autopsy. All gastrointestinal tract symptoms associated with NSDD were clearly described, including the watery content, dilatation, impaction, congestion, and mucus. On the basis of clinical signs and gastrointestinal tract symptoms, 41 rabbits were successfully sampled and classified into four groups (T1–T4) characterized by different disease severity, which would indicate the host genetic variation in susceptibility to NSDD (Table 3). The individuals could be unambiguously classified into any group were excluded. After slaughter, the intestinal sacculus rotundus tissues were immediately sampled and snap-frozen in liquid nitrogen for total RNA extraction. The 41 rabbits were also subjected to genotyping and mRNA expression analysis.
Mutational analysis of NLRP12 gene
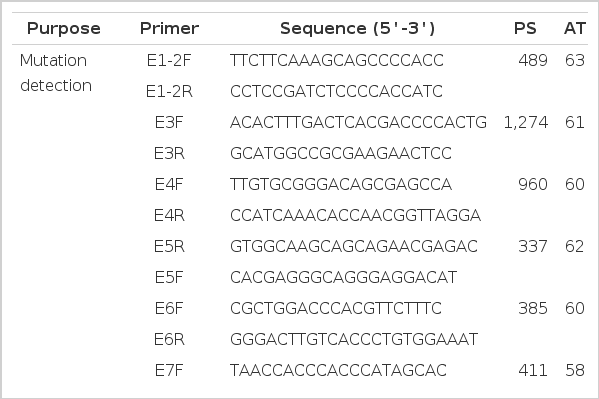
A total of 24 unrelated rabbits were randomly selected from both case and control groups for mutational analysis by sequencing the genomic region encompassing the entire NLRP12 gene. Eight PCR primer pairs were designed to amplify the eleven exons according to the reference sequence of rabbit NLRP12 gene (GenBank ID: NW_003159550) (Table 1). PCR reaction was performed in a 50 μl mixture with the following conditions: one denaturation cycle at 94°C for 5 min, 35 cycles at 94°C for 30 s, 58 to 63°C for 30 s and 72°C for 60 s, and followed by an extension cycle at 72°C for 10 min. The 50 μl reaction volume included 25 μl 2×Taq PCR MasterMix (TIANGEN, Beijing, China), 2 μl of each primer (10 pmol/μl), 5 μl DNA template (20 ng/μl) and 16 μl ddH2O. PCR products were directly sequenced using BigDye Terminator sequencing kit (Applied Biosystems, Foster City, California, USA). Sequencing was performed on a 3700 DNA sequencer (Applied Biosystems) according to the manufacturer’s instructions. The presence of mutation was confirmed by forward and reverse sequencing. For the non-synonymous mutation, we used data from PANTHER (Thomas et al., 2003) to predict the functional consequence of each of the NLRP12 variants found in the direct sequencing.
Genotyping using single-strand conformation polymorphism (SSCP)
For the coding SNP of interest (c.1682A>G), SSCP method was applied for genotyping analysis. In order to amplify the target region containing c.1682A>G, One primer pair (SSCP-F and SSCP-R, Table 1) was designed to amplify 94 bp fragments by Primer5.0 software. The PCR reaction was performed with the following conditions: one denaturation cycle at 94°C for 5 min, followed by 39 cycles at 94°C for 10 s, 59°C for 30 s, and ended with an extension cycle at 72°C for 10 min. The 10 μl reaction volume included 5 μl 2×Taq PCR MasterMix (TIANGEN, Beijing, China), 1 μl genome DNA template (20 ng/μl), 3.2 μl ddH2O, and 0.4 μl of each primer (10 pmol/μl). PCR products were electrophoresed on 1% agrose gels using 1×TBE buffer (89 mmol/L Tris, 89 mmol/L boric acid, 2 mmol/L Na2EDTA), containing 200 ng/ml GelRedTM nucleic acid gel strain. Aliquots of 5 μl PCR products were mixed with 5 μl denaturing solution (95% formamide, 25 mmol/L EDTA, 0.025% xylene-cyanole, and 0.025% bromophenol blue), heated for 10 min at 98°C and chilled on ice immediately. Denatured DNA was subjected to 12% polyacrylamide gel electrophoresis (PAGE) (190×150×1 mm) in 1× TBE buffer and constant voltage (180 V) overnight. Three known genotype (AA, AG and GG) samples by direct sequencing were added to every PAGE as reference. The gel was stained with 0.1% silver nitrate.
Quantitative real-time RT-PCR
Total RNA was extracted using the sacculus rotundus tissues from induced NSDD group by RNAiso Pure RNA Isolation Kit (TaKaRa, China). RNA was further subjected to a quality test according to the A260/280 absorbance ratio (1.6 to 1.8) and integrity of 18 S and 28 S rRNA bands on 1% formaldehyde agarose gel. Following DNase treatment, cDNA was reversely transcribed with PrimeScript RT reagent/gDNA eraser kit (TakaRa, Dalian, China) according to manufacturer’s instruction. Real-time PCR was performed by using iQ SYBR Green Supermix kit (Bio-Rad) on Bio-Rad CFX96 system following manufacturer’s instruction.
Hypoxanthine guanine phosphoribosyltransferase (HPRT, GenBank ID: NM_001105671.1) was adopted in the present study as the reference gene. The primer pairs for NLRP12 (NLRP12-qF and NLRP12-qR) and HPRT (HPRT-F and HPRT-R) genes were designed for quantitative real-time RT-PCR analysis (Table 1). RT-PCR reaction was conducted in a volume of 10 μl per reaction, including 1 μl of template cDNA (20 ng/μl), 0.3 μl of each primer, 5 μl 1×iQ SYBR Green SuperMix (Bio-Rad), and 3.4 μl ddH2O. Reaction condition was 98°C for 3 min, 40 cycles of 98°C for 5 s and 59°C for 25 s, subsequently 95°C for 10 s. In order to verify specific amplification, amplicons were systematically checked by melting curve analysis with increments of 0.5°C per cycle from 55°C to 95°C. Standard curves were generated using 10−3 to 10−10 dilution series template of PCR product for the NLRP12 and HPRT genes. Each sample was assayed in triplicate and mean values from the triplicates were used for all analyses.
Statistical analysis
DNA sequences of NLRP12 gene were assembled and aligned for mutational analysis in DNAstar program (DNAS Inc, Madison, WI, USA). HWE and frequency differences of c.1682A>G variant were tested in case and control groups. Estimates of odds ratio (OR) accompanying 95% confidence intervals (CIs) were presented for NSDD risk factor. Five inheritance models (co-dominant, dominant, recessive, over-dominant, and additive) were tested for three genotypes according to the Akaike information criteria (AIC) and Bayesian information criteria (BIC) by using SNPStats tool (Solé et al., 2006).
Because of similar PCR efficiencies (NLRP12, 99.7%; HPRT, 95.5%), gene expression was analyzed by the 2−ΔΔCT method (Livak and Schmittgen, 2001). The differences of NLRP12 mRNA expression among three genotypes (AA, AG, and GG) and four groups (T1-T4) were tested by one-way ANOVA, which followed by Tukey’s multiple comparison test. The ANOVA analysis was carried out by using GraphPad Prism 5.0 (San Diego, CA, USA). Statistical significance was set at p<0.05.
RESULTS
Mutational analysis of NLRP12 gene
We successfully amplified and sequenced the coding region of the NLRP12 gene in the rabbit. A total of 19 SNPs were identified by direct sequencing and all of them distributed over 7 exons region (Figure 1). Among them, five SNPs (c.223C>T, c.1279A>G, c.1682A>G, c.2267G>A, c.2800T>G) resulted in amino acid changes, which were p.Arg75Cys, p.Thr427Ala, p.His561Arg, p.Ser756Asn, p.Ser934Ala, respectively (Table 2). Both p.Thr427Ala and p.His561Arg were located in the exon3 which encoded the nucleotide-binding domain (NBD) domain. Another 14 SNPs were synonymous mutations.
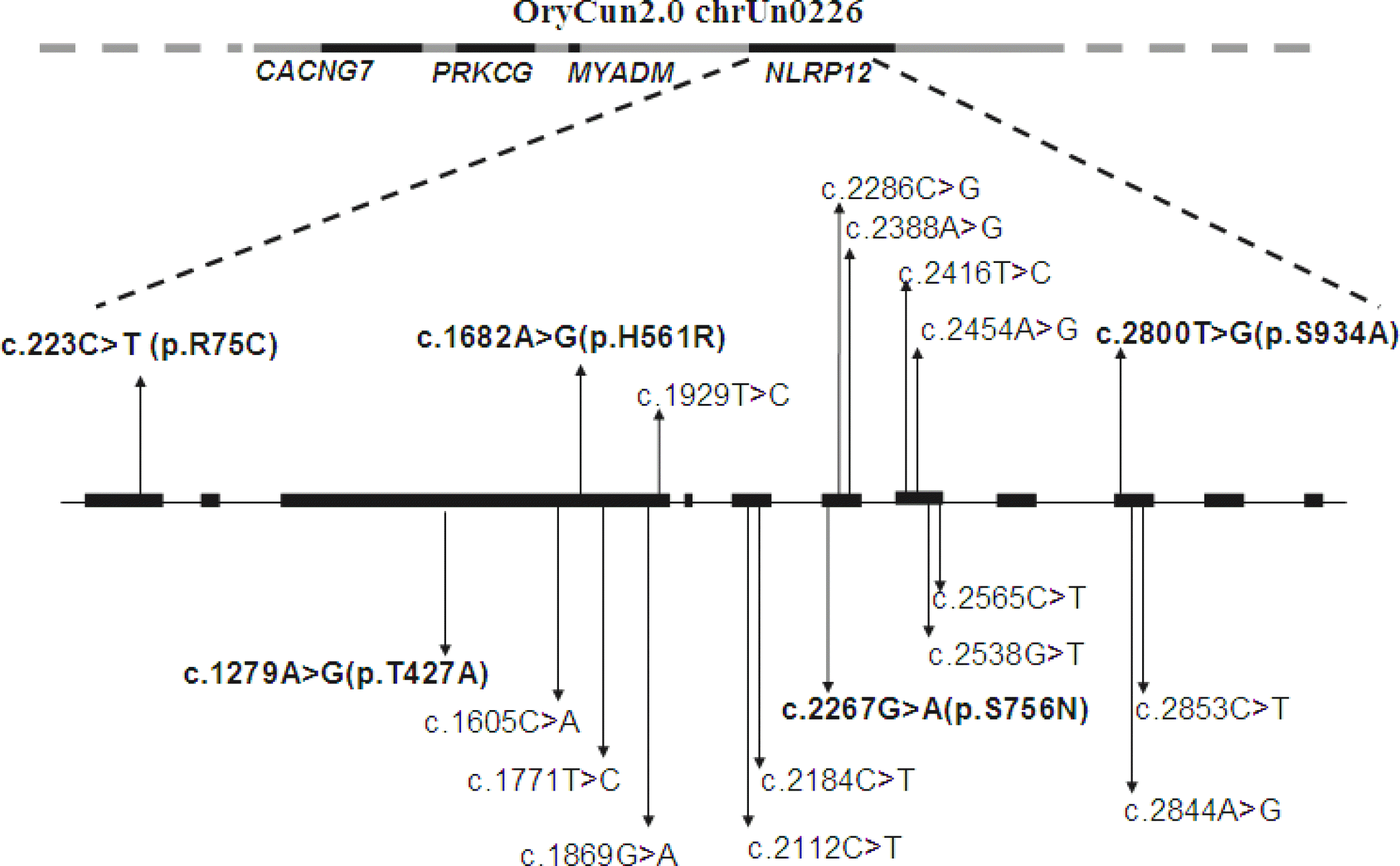
Schematic illustration of rabbit NLRP12 gene characteristic and the identified genetic variants. The relative chromosome positioning of NLRP12 gene was indicated by mapping to GeneBank Scaffold OryCun2.0 chrUn0226. The inverted arrows indicate location of identified variants in the present study, which are numbered by mRNA positions starting from translation initiation codon. Black bars represent the eleven exons. Five variants printed in bold represent non-synonymous mutation, other indicate the synonymous mutation.
Functional analysis of the missense mutations in silico
Among the five detected missense mutations, in silico functional analysis of missense mutations revealed that the subPSEC values of the five SNPs were all under -3 (Table 2). In order to search the key SNP responsible for the NSDD in rabbit, we selected c.1682A>G SNP with the lowest subPSEC value (–5.6218) for the case-control study.
Case-control association study of c.1682A>G variant
The non-synonymous SNP c.1682A>G was further subjected to genotype analysis and case-control association study (Table 4). The average frequencies of genotypes AA, AG, and GG among 539 case and control subjects were 25.6%, 52.1%, and 22.3%, respectively. The frequency of allele A in control group was significantly higher than that in case group (54.9% vs 48.5%, p<0.05). Significant deviations from Hardy-Weinberg equilibrium (HWE) were observed in control group (p = 0.026) but not in case group (p = 0.47).
Case-control association study revealed that the allele A was significantly protective for digestive disorder with an OR value of 0.88 [95% CI (confidence interval), 0.78 to 0.99; p<0.05] (Table 4). Among the five inheritance models used to evaluate importance of associated genotype, recessive inheritance was revealed as the best fit model according to the lowest values of AIC (742.1) and BIC (750.7) indexes. Under recessive inheritance model, the genotype GG was associated with increasing risk of NSDD (OR value, 1.88; 95% CI, 1.24 to 2.85), furthermore this correlation was statistically significant (p = 0.0027).
NLRP12 mRNA expression analysis
By experimentally inducing the NSDD, 41 rabbits were successfully classified into four groups (T1 to T4) characterized by different levels of genetic risk according to clinical signs and gastrointestinal tract symptoms. Through genotype analysis, the number of genotype AA, AG, and GG of c.1682A>G were 12, 18, and 11, respectively.
Among these four symptom groups (Figure 2A), the mean level of NLRP12 mRNA expression gradually decreased with the aggravation of NSDD, with the highest expression in the healthy group (T1) and lowest expression in the severe NSDD group (T4). The expression difference among different health status reached statistical significance (p = 0.0015). NLRP12 mRNA expression among the three genotypes, significant difference was found between AA and GG (Figure 2B, p<0.05), the same result also existed between AG and GG (Figure 2B, p<0.05). However, the expression difference between genotype AA and AG was not significant. These expression results intensified the indication that the allele A is protective for NSDD.
DISCUSSION
NLRP12 was first found through searching the High Throughout Genome data base of genomic sequences and was identified as a novel PYPAF (Pyrin-containing Apaf1-like proteins) family member in humans (Wang et al., 2002). In previous studies, NLRP12 has been shown to involve in both canonical and non-canonical NF-κB signaling pathways and classical and nonclassical MHC class I gene expression in vitro (Williams et al., 2003; Williams et al., 2005; Lich et al., 2007). NLRP12 could influence transmission capacity of the dendritic cell (DC) to lymph nodes, which affects the immune response in contact hypersensitivity (Arthur et al., 2010). In the present study, we found significant association between NLRP12 cSNP c.1682A>G polymorphism and NSDD in growing rabbits. This indicated that the NLRP12 protein may involve in the inflammatory signaling pathway during the NSDD process in rabbits. SNP c.1682A>G was located in the exon3 of the NLRP12 gene, which encodes the NBD domain that plays a crucial role for NLRP12 function (Ye et al., 2008). Although many coding SNPs were found in the locus of NLRP12, the c.1682A>G mutation had the highest risk in silico functional analysis. The substitution score (subPSEC) is the negative logarithm of the probability ratio of the two variant amino acids arising from a coding SNP. SubPSEC values range from 0 to −10, where 0 implies a very conservative change (unlikely to influence protein function), and more negative scores are increasingly hazardous (Thomas and Kejariwal, 2004). The association analysis also revealed that allele A was significantly associated with decreased risk of NSDD in rabbit (p<0.05). Further investigation is needed to determine the precise molecular mechanism mediating the inflammatory signaling pathway in the process of NSDD in rabbit.
The expression of NLRP12 is characterized by its predominate presence in cells of the myeloid and monocytic lineage serving as the first defense line against microorganisms (Wang et al., 2002; Williams et al., 2003). NLRP12 is a negative regulator against TLR pathway mediating up-regulation inflammation and antimicrobial response (Williams et al., 2005). So we predicted that, when NSDD occurs, the activation of TLR signaling by microorganisms may influence the expression level of NLRP12. The results of the NLRP12 expression analysis are in parallel with what we predicted. The NLRP12 mRNA level among different levels of disease severity was statistically significant different (p<0.01). A similar result was found among different NLRP12 genotypes. The result of this study was the same as that of an previous in vitro study, in which the NLRP12 expression is down-regulated after TLR activation (Williams et al., 2005). The secretion amount of IL-1β was an important hallmark indicating the severity of the inflammation in many diseases. A recent paper reported that NLRP12 silencing enhanced IL-1β release in THP-1 cells (Khare et al., 2012).
Many mutations in NLR gene are associated with inflammatory bowel diseases in human and farm animals. Mutations in NOD2 were found in patients with Crohn’s disease, Blau syndrome and in rabbit with NSDD (Hugot et al., 2001; Ogura et al., 2001; Zhang et al., 2013). Excluding NLRP12 and NOD2, other NLR members also were involved in the complicated signaling pathways mediating the cytokines and adjusting the process of inflammatory diseases. A latest study showed that NLRP6 shaped the characteristic of intestinal flora and that the loss of NLRP6 induced more colitogenic microbiota to flourish (Elinav et al., 2011). Recently, NLRC4−/− mice were found to be more impressionable to DSS-colitis, characterized with accelerated weight loss, tissue inflammation and increased cytokines (Carvalho et al., 2012). We predicted that the NLRs serve as a safety cutoff that controls the sophisticated inflammatory pathways in the gut immunological system. In conclusion, under tight surveillance of NLR family, the gastrointestinal tract could avoid potentially pro-inflammatory enteric microbial pathogens invading the host.
Deviations from Hardy-Weinberg equilibrium (HWE) can indicate inbreeding, population stratification, and even problems in genotyping (Wigginton et al., 2005). In the present study, we found deviation from HWE for c.1682A>G variant in the control group, but not in the case group. The result indicated that the genotype difference existed between case and control groups. However, the significant deviation from HWE in control group observed in the present study could not the consequence of genotyping errors because we performed strict quality control for SSCP analysis. PCR-SSCP was a simple and sensitive method for SNP detection without false-positive results and it was used widely in clinical DNA diagnosis and group analysis (Hayashi, 1991; Hayashi, 1992).
In conclusion, both the genetic association study and gene expression analysis suggested that NLRP12 participated in the manipulative process during pathogenesis of NSDD in rabbits. Disease resistance breeding has attracted increasing attention in farm animal breeding globally, so the NLRP12 gene could display predomination in the progress of molecular breeding in farm animal with NSDD. However, to obtain a convincing conclusion there needs to be further investigations, such as applying knock-out animal model and other advanced biotechnology.
Acknowledgements
This work was financially supported by the projects in the Sichuan province science and technology pillar program during the twelfth five-year plan period (2011NZ0099-4) and the earmarked fund for China agriculture research system (Grant No: CARS-44-A-2).