Efficient transgene expression system using a cumate-inducible promoter and Cre-loxP recombination in avian cells
Article information
Abstract
Objective
Transgenic technology is widely used for industrial applications and basic research. Systems that allow for genetic modification play a crucial role in biotechnology for a number of purposes, including the functional analysis of specific genes and the production of exogenous proteins. In this study, we examined and verified the cumate-inducible transgene expression system in chicken DF1 and quail QM7 cells, as well as loxP element-mediated transgene recombination using Cre recombinase in DF1 cells.
Methods
After stable transfer of the transgene with piggyBac transposon and transposase, transgene expression was induced by an appropriate concentration of cumate. Additionally, we showed that the transgene can be replaced with additional transgenes by co-transfection with the Cre recombinase expression vector.
Results
In the cumate-GFP DF1 and QM7 cells, green fluorescent protein (GFP) expression was repressed in the off state in the absence of cumate, and the GFP transgene expression was successfully induced in the presence of cumate. In the cumate-MyoD DF1 cells, MyoD transgene expression was induced by cumate, and the genes controlled by MyoD were upregulated according to the number of days in culture. Additionally, for the translocation experiments, a stable enhanced green fluorescent protein (eGFP)-expressing DF1 cell line transfected with the loxP66-eGFP-loxP71 vector was established, and DsRed-positive and eGFP-negative cells were observed after 14 days of co-transfection with the DsRed transgene and Cre recombinase indicating that the eGFP transgene was excised, and the DsRed transgene was replaced by Cre recombination.
Conclusion
Transgene induction or replacement cassette systems in avian cells can be applied in functional genomics studies of specific genes and adapted further for efficient generation of transgenic poultry to modulate target gene expression.
INTRODUCTION
Combined with the announcement and public access of the genome sequence project, genomic modification tools provide a wealth of multidisciplinary information for functional genomics. To examine the specific function(s) of genes of interest or to investigate regulatory networks, loss or gain of function studies are useful both in vitro and in vivo. Recently developed transgene delivery techniques and genome editing tools will advance biotechnology research areas much more rapidly. Many reports have demonstrated the tremendous value of genetic modification of the poultry genome for agricultural and biopharmaceutical industries [1–4].
In mammals, various transgenic inducible systems using metal ions [5] and steroid hormones [6] have been developed. However, there are limitations to these methods: transgene leakiness when expression is turned off and secondary effects from the inducers [6,7]. An alternative control element in the tetracycline resistance operon from Escherichia coli (E. coli) has been utilized to regulate transgene expression in mammalian cells [7]. A tetracycline response element consists of seven repeats of a 19-nucleotide tetracycline operator, which is recognized by a chimeric transactivator (tTA) composed of the tetracycline repressor and the VP16 activation domain [7]. Tetracycline or an analog, such as doxycycline, binds to tTA and reduces transcription of gene(s) regulated by the downstream promoter.
In both prokaryotic and eukaryotic organisms, various site-specific recombination reactions are conserved, and these systems generally involve specific enzymes and unique asymmetric DNA sequences [8–10]. Based on these versatile biological systems, Sauer and Henderson [9] developed Cre/lox-dependent site-specific recombination. Site-specific recombination technology and recombinase-mediated gene cassette exchange systems have advanced significantly and become one of the more powerful genetic manipulation tools.
Despite the various uses of inducible transgenes and the Cre/loxP system in transgenic mice, there are few reports on the production of transgenic poultry using these systems. Developmental and physiological differences have hampered development of transgenic poultry lines: adaptation of mouse-based transgenic technologies has been unsuccessful, apart from viral transduction. Recently, an efficient transgenic production system was established using piggyBac transposase and transposon elements [2,3]. In this study, we examined cumate-inducible transgene expression and loxP element-mediated transgene recombination using Cre recombinase in avian cells.
MATERIALS AND METHODS
Construction of transgene expression vectors
To construct the cumate-inducible transgene expression vector, the green fluorescent protein (GFP) or MyoD gene was inserted into the piggyBac cumate switch inducible vector (System Biosciences, Palo Alto, CA, USA) after NheI/NotI digestion and ligation. The cumate repressor, the CymR gene, was designed as an all-in-one vector system and inserted into the same piggyBac cumate-GFP and -MyoD inducible vectors. For construction of the translocation transgene expression vectors, loxP66 (5′-ata act tcg tat agc ata cat tat acg aac ggt a-3′) and loxP71 (5′-tac cgt tcg tat agc ata cat tat acg aag tta t-3′) were used for the 5′- and 3′-flanking regions, respectively. Firstly, the transgene expression vector carrying the cytomegalovirus-enhanced green fluorescent protein (CMV-eGFP) gene was inserted into the 5′- and 3′-UTR piggyBac transposon elements (piggyBac loxP66-eGFP-loxP71). For the translocation vectors, CMV-DsRed (loxP66-DsRed-loxP71) was inserted between the loxP66 and loxP71 sequences. The promoter of elongation factor 1 was used for expression of the puromycin-resistance gene in the transgene expression vectors, and the CMV promoter was used for expression of the Cre recombinase gene (Addgene; www.addgene.org).
Cell culture and transfection
Chicken DF1 cells (American Type Culture Collection [ATCC], Manassas, VA, USA) were maintained and passaged in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (FBS; Invitrogen, USA) and 1× antibiotic-antimycotic (Invitrogen, USA). These cells were cultured in an incubator at 37°C in an atmosphere of 5% CO2 and 60% to 70% relative humidity. QM7 cells (ATCC) were maintained under the same conditions in Medium 199 containing 10% FBS (HyClone, South Logan, UT, USA), 2% chicken serum (Sigma-Aldrich, St. Louis, MO, USA), 1×antibiotic-antimycotic (Invitrogen, USA) and subcultured at 70% confluency. To establish the transgene-expressing cell lines, each transgene expression vector was co-transfected with piggyBac transposase using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s protocol. Once the seeded cells reached 80% confluency on 6-well culture plates, they were washed with phosphate-buffered saline (PBS) and refreshed with 2 mL of the culture media without antibiotic-antimycotic. The plasmid DNA-lipid complex consisting of 7.5 μL Lipofectamine 3000 reagent in 250 μL Opti-MEM (Invitrogen, USA) and 10 μL P3000 reagent with 2.5 μg piggyBac transgene vector and piggyBac transposase plasmid in 250 μL Opti-MEM was added to each well. One day after lipofection, 10 μg puromycin/mL was added to select the sublines stably transfected with the transgene. To induce transgene expression, the cumate solution (System Biosciences, USA) was added, and the culture media were changed every 3 days. For the translocation experiments, the loxP66-eGFP-loxP71 vector was co-transfected with Cre recombinase using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s protocol.
Flow cytometry
After treatment with 0.05% trypsin-ethylenediaminetetraacetic acid, the cells were resuspended in PBS containing 1% BSA and strained through a 40 μm cell strainer (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) for flow cytometry. Data analysis was performed using FlowJo software (Tree Star Inc., Ashland, OR, USA; http://www.treestar.com).
Quantitative RT-PCR analysis
Total RNA was extracted using Trizol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Total RNA was quantified using a NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA), and 2 μg RNA were used for cDNA synthesis using random primers (Invitrogen, USA) under standard conditions. Each 20 μL reverse transcription polymerase chain reaction (RT-PCR) reaction mix contained 2 μL cDNA, 2 μL PCR buffer, 1.6 μL dNTP mixture (2.5 mM), 1 unit Taq DNA polymerase, and 10 pmol forward and reverse primer (Table 1). Quantitative RT-PCR analysis was performed using the iCycler iQ Real-time PCR detection system (Bio-Rad, Hercules, CA, USA) and EvaGreen (Biotium, Fremont, CA, USA). The PCR parameters were as follows: an initial incubation at 94°C for 5 min, followed by 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The reaction was terminated by a final incubation at 72°C for 10 min, and melting curve profiles were analyzed for the amplicons.
Statistical analysis
Statistical analysis was performed using the Student’s t-test in SAS version 9.3 software (SAS Institute, Cary, NC, USA). Significant differences between the groups were determined using the general linear model (PROC-GLM) in SAS software. Differences between treatments were deemed significant when the p value was less than 0.05.
RESULTS
Cumate-inducible transgene expression
After co-transfection of the piggyBac cumate-GFP transgene vector and piggyBac transposase construct into DF1 and QM7 cells, stable cells were selected using puromycin. There were no GFP-expressing cells in the off state in the cumate-GFP DF1 cells (Figure 1A). In contrast, slightly leaky GFP expression was observed in the QM7 cells (Figure 1A). To confirm the leaky transgene expression in the off state, flow cytometric analysis was conducted in both cumate-GFP DF1 and QM7 cells (Figure 1B). Fewer than 5% of the GFP-positive cells were detected only in the cumate-GFP QM7 cells (Figure 1B). In the presence of cumate, GFP transgene expression was induced in both the cumate-GFP DF1 and QM7 cells. Moreover, GFP expression was stronger in the cultures treated with cumate for 6 days than in those cultured for 3 days (Figure 1C).
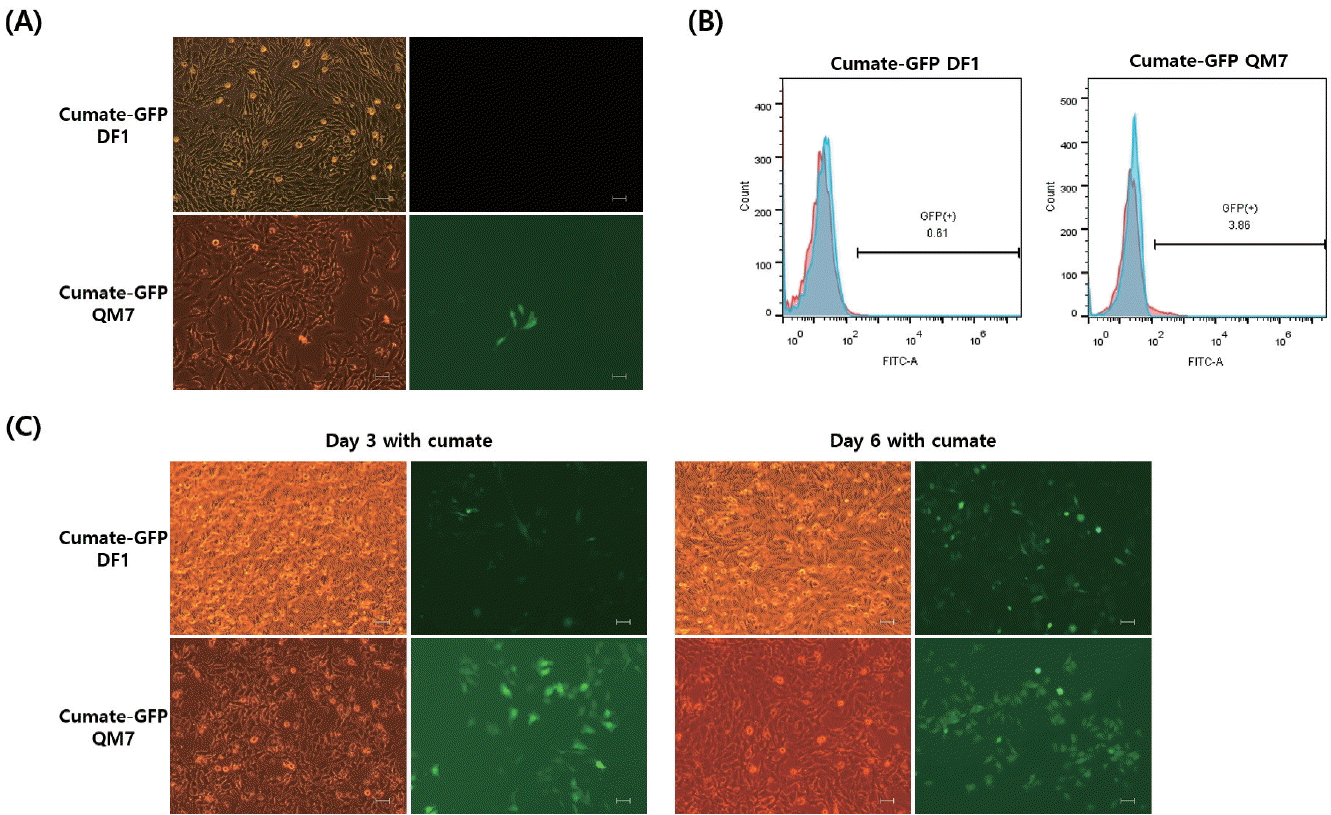
Controlled inducible transgene expression in chicken DF1 and quail QM7 cells. (A) Leaky expression of the (green fluorescent protein (GFP) transgene is regulated by the cumate promoter. In DF1 cells, there is no GFP expression in the absence of cumate (upper panel), but slightly leaky GFP expression was apparent in QM7 cells. (B) Flow cytometric analysis of GFP-expressing cells among cumate-GFP DF1 and QM7 cells in the off condition (in the absence of cumate). (C) The cumate-GFP cells were cultured in the presence of cumate for 3 or 6 days. All figures show phase contrast (left) and fluorescent (right) images (all fluorescent images were observed under the fluorescent microscope, Scale bar = 100 μm).
To examine the dose-dependent control of GFP transgene expression by cumate, flow cytometry was performed following induction with different concentrations of cumate. In the cumate-GFP cells, the percentage of GFP-expressing cells increased continually (Figure 2). In the cumate-GFP DF1 cells, 450 μg cumate/mL significantly induced GFP transgene expression compared with 300 μg/mL at 3 and 6 days, but no significant difference was found between 3 and 6 days (55.07%±5.22% and 62.37%±0.23% with 300 μg/mL at 3 and 6 days, respectively; 55.63%±1.72% and 64.23%±1.8% with 450 μg/mL at 3 and 6 days, respectively, Figure 2A). The percentage of GFP-positive cells in the cumate-GFP QM7 cells also increased depending on the concentration of cumate (Figure 2B). However, the induction efficiency of transgene expression in both 3 and 6 day cultures was much lower in the cumate-GFP QM7 cells compared with the cumate-GFP DF1 cells (Figure 2A, 2B). In the cumate-GFP QM7 cells, treatment with 450 μg cumate/mL showed slightly increased induction of the GFP transgene at 6 days compared with 3 days (10.17%±0.29% at 3 days vs 12.27%± 0.91% at 6 days, Figure 2B).
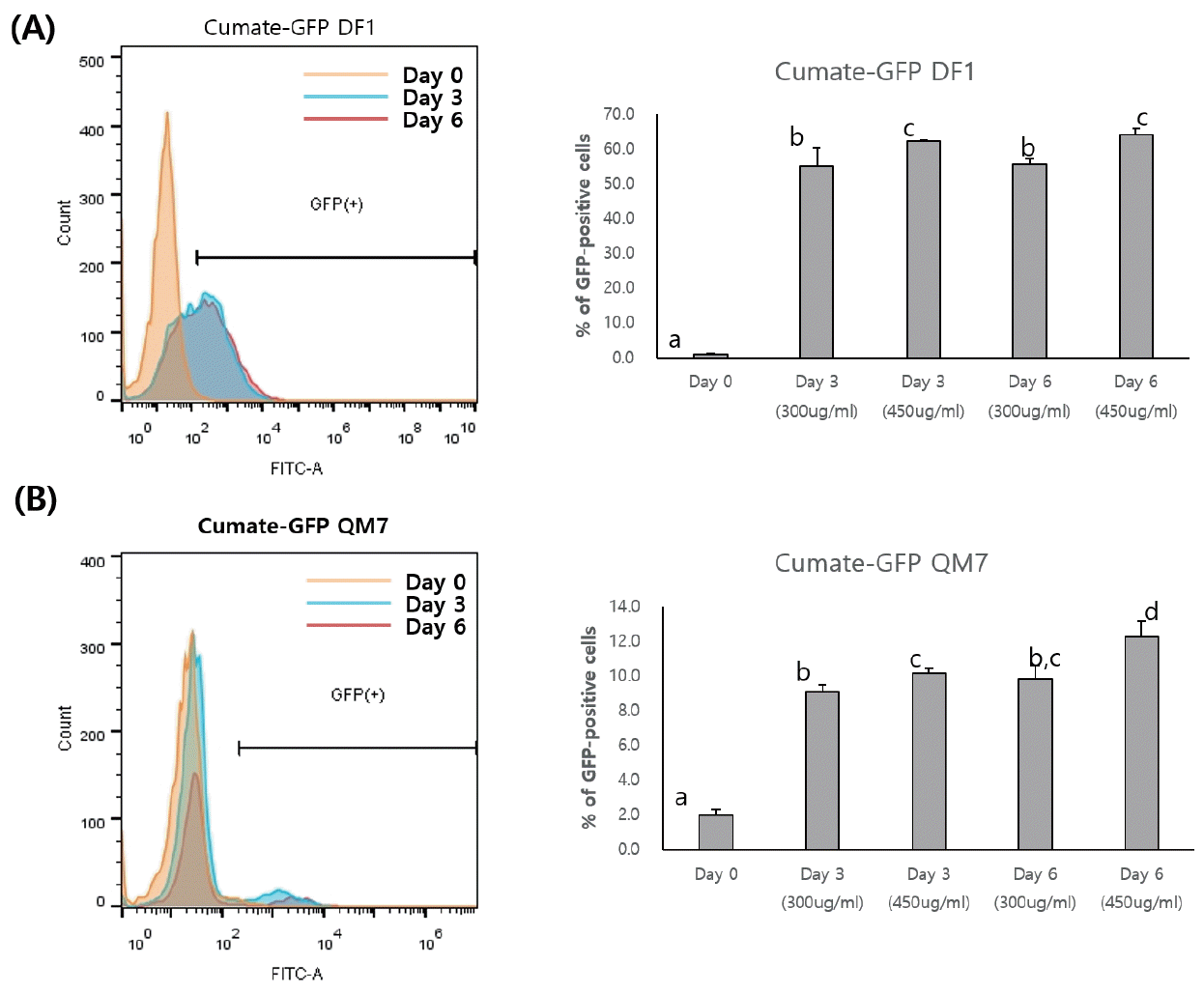
Flow cytometry following induction of the green fluorescent protein (GFP) transgene by different concentrations of cumate. Flow cytometry determination of the percentages of GFP-positive cells after cumate induction in cumate-GFP DF1 (A) and cumate-GFP QM7 (B) cells. The different superscripts next to the percentages of GFP-positive cells indicate statistically significant differences between treatments (p<0.05).
Cumate-induced MyoD gene expression and qRT-PCR of myogenic genes
To apply the cumate-inducible transgene expression system, the MyoD gene, an important regulator of muscle differentiation, was inserted into the piggyBac cumate-inducible expression vector, and stably transfected DF1 cells were established following transfection and puromycin selection. After induction of the cumate-MyoD DF1 cells with 450 μg cumate/mL, the levels of myogenic genes, in particular myogenin and desmin, were determined by quantitative RT-PCR. MyoD gene expression increased continually depending on the length of exposure to the cumate solution (Figure 3A). Similarly, myogenin and desmin expression were also significantly upregulated after exposure to cumate (Figure 3B, 3C). These results suggest that the cumate-inducible transgene expression platform can be efficiently utilized to investigate the biofunction(s) of specific genes in an in vitro cell culture system.
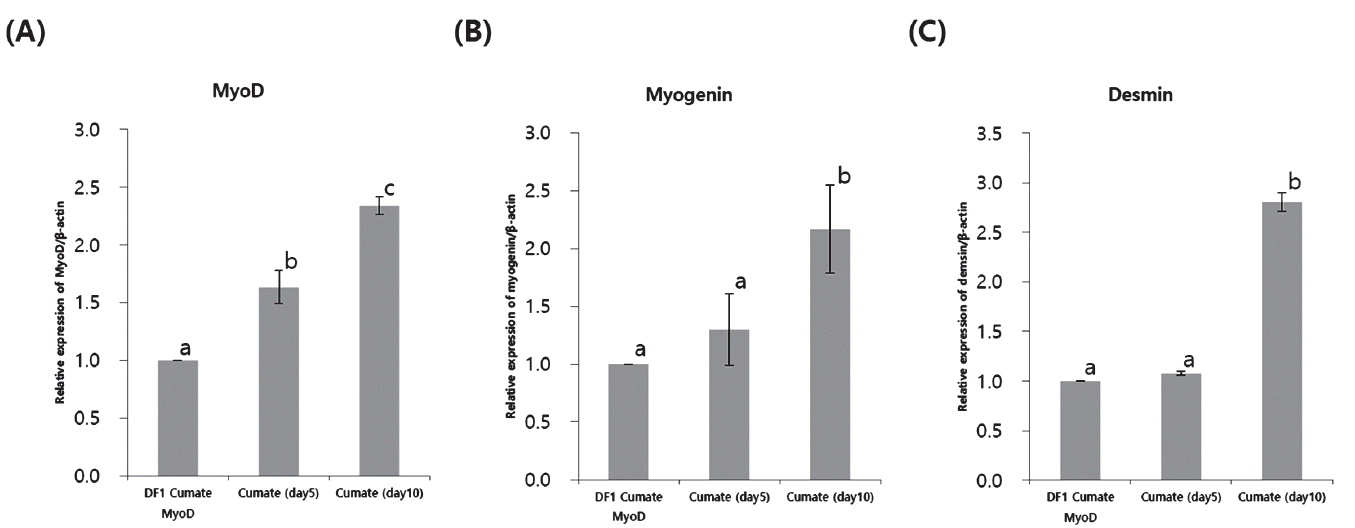
Quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis in cumate-MyoD DF1 cells following cumate induction. The expression of MyoD. (A), myogenin (B), and desmin (C) are expressed relative to β-actin expression. The different superscripts of the relative expression for each transcript indicate statistically significant differences between treatments (p<0.05).
Cre recombinase-mediated transgene translocation
For the translocation experiments, a stable eGFP-expressing DF1 cell line transfected with the loxP66-eGFP-loxP71 vector (Figure 4A) was established, and the loxP66-DsRed-loxP71 transgene vector was co-transfected with Cre recombinase. After 14 days of co-transfection, DsRed-positive and eGFP-negative cells were observed, indicating that the eGFP transgene was excised, and the DsRed transgene was replaced by Cre recombination (Figure 4B).
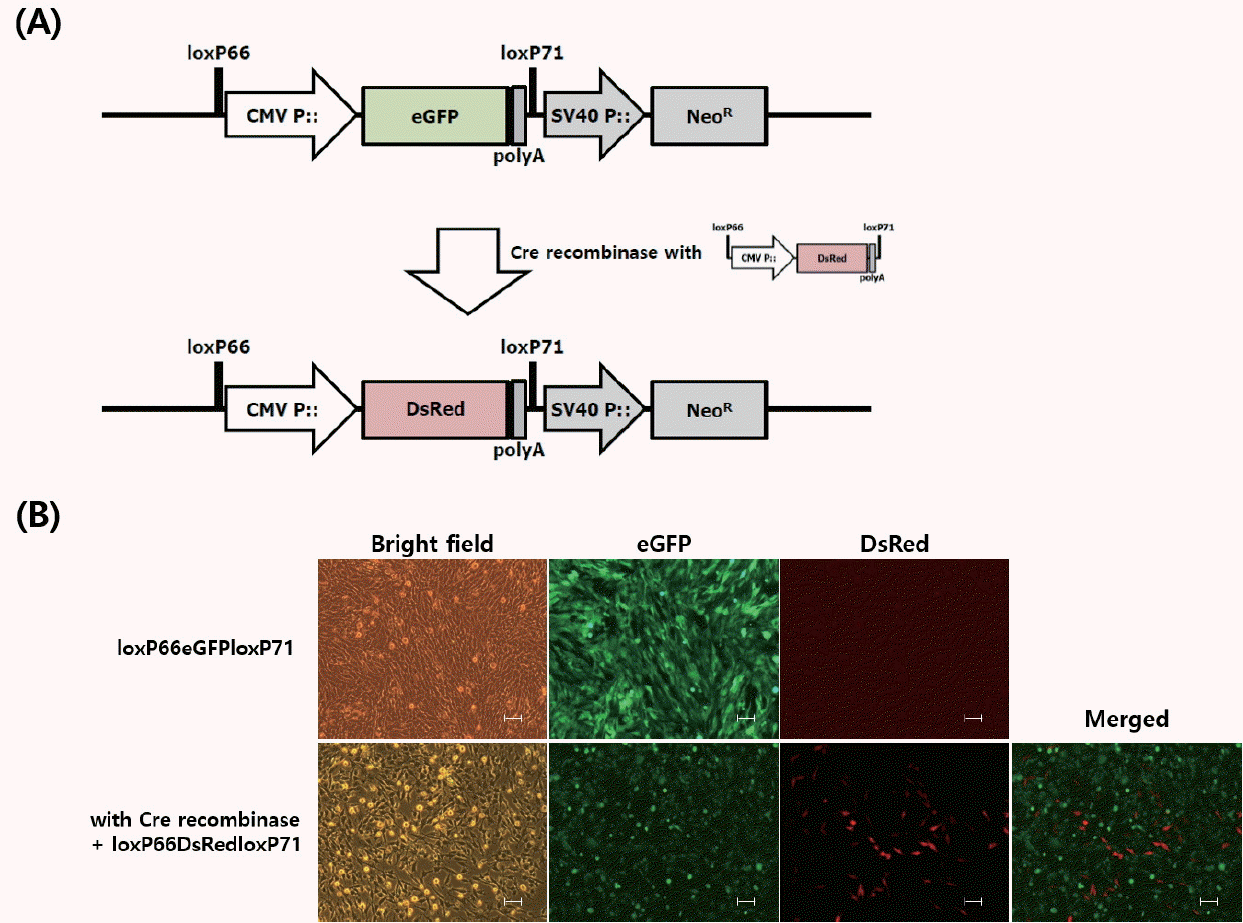
Transgene translocation analysis in loxP66-eGFP-loxP71 expression vector-transfected DF1 cells after co-transfection with the Cre recombinase expression and loxP66-DsRed-loxP71 vectors. (A) Schematic of the expression vector used for transgene translocation with Cre recombinase. loxP66 and loxP71 sequences were used for Cre recombinase recognition, and the cytomegalovirus (CMV) promoter was designed for transgene expression. The enhanced green fluorescent protein (eGFP) transgene was replaced with the DsRed transgene following Cre recombination. (B) Translocation analysis of transgene expression in loxP66-eGFP-loxP71 expression vector-transfected cells after 14 d of co-transfection with the Cre recombinase expression and loxP66-DsRed-loxP71 vectors. After 14 d of co-transfection, positive cells were identified as cells in which the expression of eGFP and DsRed did not overlap, indicating that the eGFP transgene was excised, and the DsRed transgene was replaced by Cre recombination (all fluorescent images were observed under the fluorescent microscope, Scale bar = 100 μm).
DISCUSSION
To advance the study of functional gene activity and regulatory processes, a tightly controlled inducible expression system of specific genes will be an important prerequisite platform. Various inducible transgene systems have been developed [7,10], including the generation of transgenic chickens by retrovirus-mediated tetracycline-inducible expression vector delivery [11]. A new technical platform, the cumate-inducible system, has recently been designed and developed for the efficient induction of transgene expression [12]. Similar to the tetracycline induction system, the CymR repressor binds cumate operator sequences and inhibits transgene expression by suppressing the downstream promoter. This repression can be alleviated by the addition of cumate, a non-toxic molecule that binds to CymR [12]. This cumate-controlled system shows dynamic transgene expression inducibility that can be finely tuned and is reversible in the absence of cumate [12]. Thus, we have examined a new system for transgene expression in avian cell lines. The transfer of the piggyBac transposon-mediated transgene was efficient, and the transgenic avian cell lines were stably established following puromycin selection. In cumate-GFP DF1 cells, GFP expression was repressed in the off state in the absence of cumate (Figure 1). However, cumate-GFP QM7 cells exhibited low expression of the GFP transgene in the absence of cumate, indicating that control of transgene expression using the cumate-inducible promoter differs among different types of avian cells (Figure 1). After culture in the presence of cumate, transgene induction efficiency in the cumate-GFP DF1 cells was higher than that in the cumate-GFP QM7 cells regardless of the cumate concentration or the number of days in culture (Figure 2). Additionally, cumate-GFP QM7 cells exhibited cellular toxicity at both concentrations of cumate, whereas the cumate-GFP DF1 cells were maintained for a long period in the presence of cumate. This result suggests that the cumate-inducible system is suitable for chicken DF1 cells. Following this, we constructed a cumate-regulated MyoD expression vector and induced transgene expression in DF1 cells to examine the functional activity of MyoD. In cumate-MyoD DF1 cells, MyoD transgene expression was successfully induced by cumate, and the genes controlled by MyoD were upregulated according to the number of days in culture. This inducible expression system of myogenic genes could be utilized to investigate the mechanisms underlying muscle differentiation.
The versatile Cre/loxP recombination system is commonly used for genetic manipulation of mammalian cell lines and transgenic animals [9,10]. This site-specific recombination technology is widely used to delete, insert, translocate, invert, and exchange a gene of interest at a target site [9,10]. The 34-nucleotide loxP sequences were originally derived from bacteriophage P1 [8]. loxP elements include asymmetric 8-nucleotide spacer sequences, which are variable except for the middle two bases positioned between two sets of 13-nucleotide palindromic sequences [8]. Due to the inefficiency of wild-type loxP, mutant loxP elements, including loxP66 and loxP71, have been developed to improve the efficiency of Cre-mediated insertion or exchange [13]. In the DNA recombination reaction, a single Cre recombinase recognizes and rearranges the reciprocal sequences of target loxP elements. In particular, Cre/loxP-targeted DNA modification is useful when the mutant phenotype induced by global expression or knockout of the target gene is lethal during embryonic development or following birth [14]. The Cre/loxP recombination system efficiently regulates expression or deletion of the transgene in a spatiotemporal manner through conditional gene activation or knockout combined with a tissue-specific or inducible promoter to trigger recombination activity. In birds, however, there are relatively few reports on the application of recombinase-mediated gene cassette exchange using the chicken DT40 cell line [13,15]. In this study, we examined the cumate-inducible transgene expression system and loxP element-mediated transgene recombination using Cre recombinase in avian cell lines. These versatile technical platforms could be efficiently adapted for use in functional genomics studies and for the generation of transgenic poultry to regulate the expression of specific target genes.
ACKNOWLEDGMENTS
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01111401)” Rural Development Administration, Republic of Korea.
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.