Characterization of the Nanog 5′-flanking Region in Bovine
Article information
Abstract
Bovine embryonic stem cells have potential for use in research, such as transgenic cattle generation and the study of developmental gene regulation. The Nanog may play a critical role in maintenance of the undifferentiated state of embryonic stem cells in the bovine, as in murine and human. Nevertheless, efforts to study the bovine Nanog for pluripotency-maintaining factors have been insufficient. In this study, in order to understand the mechanisms of transcriptional regulation of the bovine Nanog, the 5′-flanking region of the Nanog was isolated from ear cells of Hanwoo. Results of transient transfection using a luciferase reporter gene under the control of serially deleted 5′-flanking sequences revealed that the −134 to −19 region contained the positive regulatory sequences for the transcription of the bovine Nanog. Results from mutagenesis studies demonstrated that the Sp1-binding site that is located in the proximal promoter region plays an important role in transcriptional activity of the bovine Nanog promoter. The electrophoretic mobility shift assay with the Sp1 specific antibody confirmed the specific binding of Sp1 transcription factor to this site. In addition, significant inhibition of Nanog promoter activity by the Sp1 mutant was observed in murine embryonic stem cells. Furthermore, chromatin-immunoprecipitation assay with the Sp1 specific antibody confirmed the specific binding of Sp1 transcription factor to this site. These results suggest that Sp1 is an essential regulatory factor for bovine Nanog transcriptional activity.
INTRODUCTION
Nanog, a homeodomain transcription factor, plays a crucial role in maintenance of the undifferentiated state of embryonic stem cells (ESCs) (Chambers et al., 2003). Its expression has been reported in ESC, embryonic carcinoma cells, and embryonic germ cells, as well as interior cells of compacted morula and epiblasts (Mitsui et al., 2003). In addition, it’s down regulation in implantation stage embryos and disappearance from the expression profile along with differentiation of ESCs into other cell types have been observed (Mitsui et al., 2003). Overexpression of Nanog in ESCs renders them leukemia inhibitory factor (LIF)-signal transducer and activator of transcription 3 (STAT3) independent for self-renewal and pluripotency. Nonetheless, cooperation of Nanog with other key pluripotent factors, such as POU domain-containing octamer-binding transcription factor 4 (Oct4) and high-mobility group (HMG)-containing sex determining region Y-box 2 (Sox2), for control of a set of target genes required for maintenance of ESC pluripotency has been reported (Pan and Thomson, 2007).
In the previous study, the Nanog and its promoter region have been isolated, sequenced, and characterized for elucidation of their functions in murine and human (Hart et al., 2004). In human, transcriptional activity analyses of the Nanog promoter revealed the presence of several cis-regulatory elements, e.g., the Oct4/Sox2 binding site and a Smad-binding element (Xu et al., 2008). Both Oct4 and Sox2 play a crucial role in transcriptional activation of Nanog expression in both murine and human ESC (Kuroda et al., 2005; Rodda et al., 2005). The promoter region of the mouse Nanog was found to contain two binding sites, one for Krüppel-like-zinc-finger transcription factor (KLF) and one for Sp1, which are indispensable for activation of the Nanog promoter (Wu and Yao, 2006). In addition, KLF4, a member of the KLF family, increased transcriptional activity of the Nanog promoter in cooperation with Oct4 and Sox2 (Wei et al., 2009) or with a homeodomain transcription factor PBX1 in human ESC (Chan et al., 2009).
It is reasonable to assume that the mechanisms that regulate the stemness of ESC might be conserved across species, from the mouse to other mammalian species, such as bovine and porcine. However, previous studies have described striking differences in distribution of stemness genes between human, mouse, and bovine embryos. Expression of Nanog has been observed in both the inner cell mass (ICM) and trophectoderm of bovine blastocytes; however, its expression is restricted to the ICM in human and mouse blastocytes (Chambers et al., 2003; Mitsui et al., 2003; Kuijk et al., 2008; Munoz et al., 2008). In addition, despite of the significant similarity of stem cell markers expression between humane and murine ESC, they possess their own distinct gene expression signatures, which may reflect species-specific differences, not culture conditions (Ginis et al., 2004; Kuijk et al., 2008; Xie et al., 2010). In this regard, care should be taken in the effort to employ the approaches to derive the ESC from one species to another (Keefer et al., 2007). No study to address this matter in other mammalian species, e.g., bovine, porcine, and ovine, has been conducted.
Therefore, in the present study, we isolated the bovine Nanog proximal promoter sequences which presumably contain evolutionarily conserved cis-regulatory sequences, including Oct4/Sox2, NF-κB, and Sp1 binding sites, and performed experiments in order to determine their roles in transcriptional activation of the bovine Nanog expression. Results from promoter assay, electrophoretic mobility shift assay (EMSA) and chromatin-immunoprecipitation (ChIP) assay revealed that Sp1 transcription factor regulates the bovine Nanog proximal promoter activity through direct binding to a Sp1-binding site in the bovine Nanog proximal promoter, whereas NF-κB, which plays an important role in transcriptional activation of the Nanog expression in both human and murine ESC, is dispensable in the bovine Nanog promoter activity.
MATERIALS AND METHODS
Isolation of the 5′-flanking region of the bovine Nanog
To isolate the 5′-flanking region of the bovine Nanog, bovine genomic DNA was isolated from cells of Hanwoo (Korean cattle) using the Wizard Genomic DNA purification kit (Promega, Madison, WI, USA) and used as template DNA for polymerase chain reaction (PCR) amplification. Gene-specific primers were used for PCR amplification to obtain a 3,094 bp-upstream DNA fragment containing the ATG translation start site of the bovine Nanog (Table 1). PCR amplification was performed in a 50 μL reaction mixture containing 1 μL of bovine genomic DNA, 20 pmol of each of the primers, 0.4 mM dNTPs, 3 mM MgCl2, 1.25 U Taq DNA polymerase (Takara), and PCR buffer. The following PCR conditions were used: 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 64°C for 45 s, extension at 72°C for 1 min, and the final extension step was prolonged to 5 min. Using the pGEM-T easy vector system (Promega, USA), isolation and analysis of positive cloned DNA by restriction enzyme mapping was performed, followed by complete sequencing in both directions (GenBank Accession No. NM_0010253).
Construction of promoter-luciferase reporter plasmids and site-directed mutagenesis
Series of plasmid constructs containing DNA fragments of various sizes from the proximal promoter region of the bovine Nanog (−2,487/+84), (−1,702/+84), (−961/+84), (−420/+84), (−290/+84), (−188/+84), (−134/+84), and (−19/+84) were constructed by insertion of DNA fragments between the KpnI and BglII sites of the pGL3-basic vector (Promega, USA). For amplification of all plasmid DNA constructs, PCR was performed using the 3,094 bp-upstream DNA fragment containing ATG (−2,904/+190) as a template with primers (Table 1). Briefly, PCR was performed using synthetic oligonucleotides that incorporated a 5′-end KpnI site and a 3′-end BglII site to obtain 5′-end-deleted DNA fragments (underlined in Table 1). For construction of the Sp1 and NF-κB mutant constructs, the oligonucleotide sequences were replaced by introduction of the XhoI enzyme site using a method based on site-directed mutagenesis PCR with pbNanog-Luc (−134/+84) as a template. Primers are shown in Table 1, in which XhoI enzyme site is underlined. Nucleotide sequencing was performed for verification of sequences and orientations of the constructs. To construct pbNanog-EGFP, the bovine Nanog promoter DNA fragments from pbNanog-Luc (−134/+84) and pbNanog-Luc (−134/+84)-mutSp1 were replaced with human Nanog promoter sequences in recombinant plasmids containing murine and human Nanog promoter tagging-pEGFP-N1 constructs which were kindly provided by Dr. Takashi Tada, Laboratory of Stem Cell Engineering at Kyoto University (Kyoto, Japan).
Cell culture and transient transfection assay
HEK293T cells (human embryonic kidney cell line), EBTr cells (bovine trachea cell line), MDBK cells (bovine kidney cell line) were cultured for the luciferase-reporter assay. All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin (Life Technologies, Carlsbad, CA, USA). Cells were seeded at 5×104 cells per well in 24-well plates 24 h prior to transfection. The R1 cells, a mouse ESC line, was cultured on mitomycin-C (Sigma-Aldrich, St. Louis MO, USA) treated mouse primary embryonic fibroblast feeder cells and grown in DMEM (Life Technologies, USA) with 15% fetal calf serum (HyClone), 0.1 mM 2-mercaptoethanol (Life Technologies, USA), 1% non-essential amino acids (Life Technologies, USA), 2 mM L-glutamine (Life Technologies, USA), 1 mM minimum essential medium (MEM) sodium pyruvate solution (Life Technologies, USA), 1% penicillin/streptomycin (Life Technologies, USA), and 1,000 U/mL LIF (ES-GRO, Chemicon, Billerica, MA, USA). Cells were maintained at 37°C with 5% CO2.
A Qiagen tip-100 column was used for purification of plasmid DNA. For bovine cells, transient transfection of plasmids for the luciferase reporter assay was performed using 0.5 μg each of the promoter-reporter constructs with 2 μL of jetPEI (Polyplus transfection, Ilkirch, France) per μg of plasmid DNA. 0.05 μg of the pRL-TK (Promega, USA) was co-transfected for normalization of transfection efficiency. Cell extracts were prepared using 100 μL passive lysis buffer (Promega, USA) 24 h after transfection, followed by assay for luciferase activity using the dual luciferase reporter assay system according to the manufacturer’s protocols (Promega, USA). All transfections were repeated three times. Ratios of the luciferase and pRL-TK activities were calculated for normalization of data. The average luciferase activity is presented as fold activation relative to the activity of the pGL3-basic vector.
For transfection of R1 cells, bovine wild type (−134/+84) and Sp1 mutant Nanog promoter-green fluorescence protein (GFP) reporter vectors and pRL-TK, a transfection control vector, were linearized, and were electroporated into 1×107 R1 cells at 1,400 V, 10 ms, and pulse number 3 using a Neon Transfection System (Life Technologies, USA). Transcriptional activity of the bovine Nanog wild type and Sp1 mutant proximal promoters was monitored under fluorescent microscope with FITC filter. 24 h after transfection, cells were lysate with RIPA buffer and Western blot followed with GFP antibody (Agilent Technologies Genomics, Santa Clara, CA, USA). pRL-TK was measured with a luminometer and used for normalization of transfection efficiency as described above.
Preparation of nuclear extract and electrophoretic mobility shift assay
For the EMSA, nuclear extracts from bovine Nanog promoter transfected R1 cells were prepared as previously described (Lee et al., 2006). Oligonucleotides used as probes for EMSA were end-labeled with biotin using a Biotin 3′end DNA labeling kit (Pierce Biotechnology, Rockford, IL, USA). The probes (as shown in Table 1) were annealed to their respective antisense strands. The Lightshift Chemiluminescent EMSA kit (Pierce Biotechnology, USA) was used for the binding reactions; 2 μg of nuclear extract from the R1 murine ESC was incubated with binding reaction buffer, 2.5% glycerol, 5 mM MgCl2, 1 μg Poly (dI•dC), and 0.1% NP-40 in the presence or absence of competitor for 1 min at room temperature. Biotin labeled, double-stranded oligomers were added to the reaction mixture for an additional 20 min at room temperature. For the competition experiments, unlabeled competitors containing intact or mutant Sp1 consensus sequences were added at 100-fold excess. Electrophoresis of the reaction products was performed at 100 V in a 6% polyacrylamide gel in 0.5×Tris-borate-EDTA (TBE) buffer. After electrophoretic transfer to a nylon membrane at 380 mA for 30 min, the membrane was cross-linked using UV-light cross-linker, followed by analysis using a Chemiluminescent nucleic acid detection module (Pierce Biotechnology, USA) according to the manufacturer’s instructions.
Chromatin-immunoprecipitation assay with Sp1 antibody
The Chromatin-immunoprecipitation (ChIP) assay was performed by using SimpleChIP enzymatic chromatin IP kit (Cell Signaling Technology, Danvers, MA, USA) according to the manufacturer’s instruction. Briefly, nuclear lysates isolated from MDBK cells were sonicated, and cross-linked proteins were immunoprecipitated by incubation with antibody against Sp1 (Santa Cruz Biotechology Inc., Dallas, Texas, USA) as well as normal IgGs (Cell Signaling Technology, USA), respectively. A DNA sample from sonicated lysate was used as internal control (input). A DNA sample that was immunoprecipitaed by normal IgGs was used as negative control. Immunoprecipitaed DNA was detected by PCR amplification using ChIP primers (Table 1) with rTaq Plus PCR Master Mix (Elpis Biotech Inc., Daejeon, Korea) at 95°C for 5 min, 34 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 30s, completed by a 5 min incubation at 72°C and PCR products were analyzed by 2% agarose gel electrophoresis.
RESULTS
Transcriptional regulation of the bovine Nanog promoter
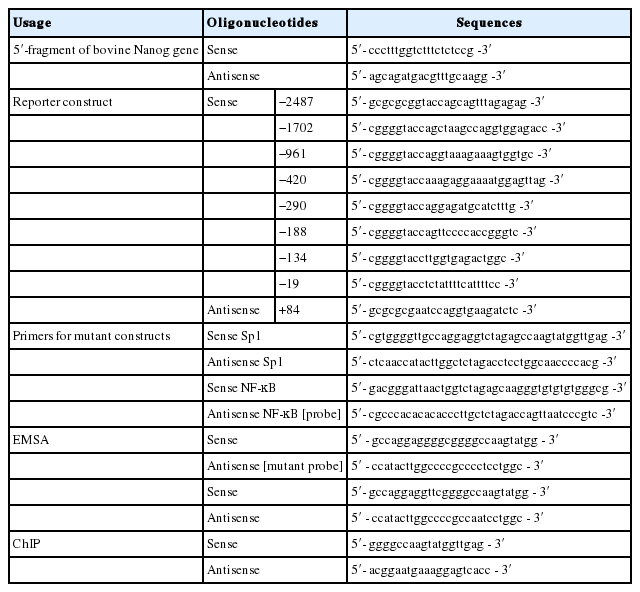
We isolated a 3,094 bp genomic fragment (−2,904/+190), including the ATG translational start codon from Hanwoo, the Korean cattle, and sequenced the isolated DNA fragment. In order to identify the conserved cis-regulatory elements involved in regulating transcription of the bovine Nanog, a series of deleted fragments of the 5′-proximal promoter region were cloned into the luciferase reporter vector (Figure 1A). Approximately 7-fold higher promoter activity was observed in the wild-type construct, i.e., pbNanog-Luc (−2,487/+84), compared with the vector control (pGL3-basic) (p<0.05). Results of deletion mutant analyses showed significant luciferase activity, compared to the vector control, and deletion between −134 and −19 resulted in abolition of transcriptional activity in HEK293T and two bovine somatic cell lines, EBTr and MDBK (Figure 1B). It is notable that transcription activity of deletion mutant pbNanog-Luc (−134/+84) was approximately 15-fold higher than that of the vector control, and more than 2-fold higher than with that of the pbNanog-Luc (−2,487/+84) construct in cell lines tested in this study (HEK293T, EBTr, and MDBK) (p<0.05, Figure 1B). In addition, pbNanog-Luc (−420/+84) also demonstrated increased transcription activity in bovine cells lines and in the HEK293T cell line (p<0.05, Figure 1B). These results indicates that the upstream regions in the bovine Nanog promoter, including the −134/+84 and −420/+84 regions, may contain positive and/or negative cis-regulatory elements involved in regulating transcription activity of the Nanog promoter.
Regulation of the bovine Nanog 5′-flanking region and promoter activities in HEK293T, EBTr, and MBDK cell lines. (A) Schematics of the series of the bovine Nanog promoter deletion construct. (B) Bovine Nanog 5′-deletion constructs and pRL-TK were transfected into HEK293T cells (human embryonic kidney cell line, top), EBTr cells (bovine trachea cell line, middle), and MDBK cells (bovine kidney cell line, bottom). Cells were harvested after 48 h, followed by measurement of luciferase activity. Firefly luciferase activity was normalized to Renilla luciferase activity. Data shown represent the mean±standard deviation of three independent experiments. HEK293T cells, human embryonic kidney cell line; EBTr cells, bovine trachea cell line; MDBK cells, bovine kidney cell line.
Requirement of the Sp1 binding site for activation of the bovine Nanog proximal promoter
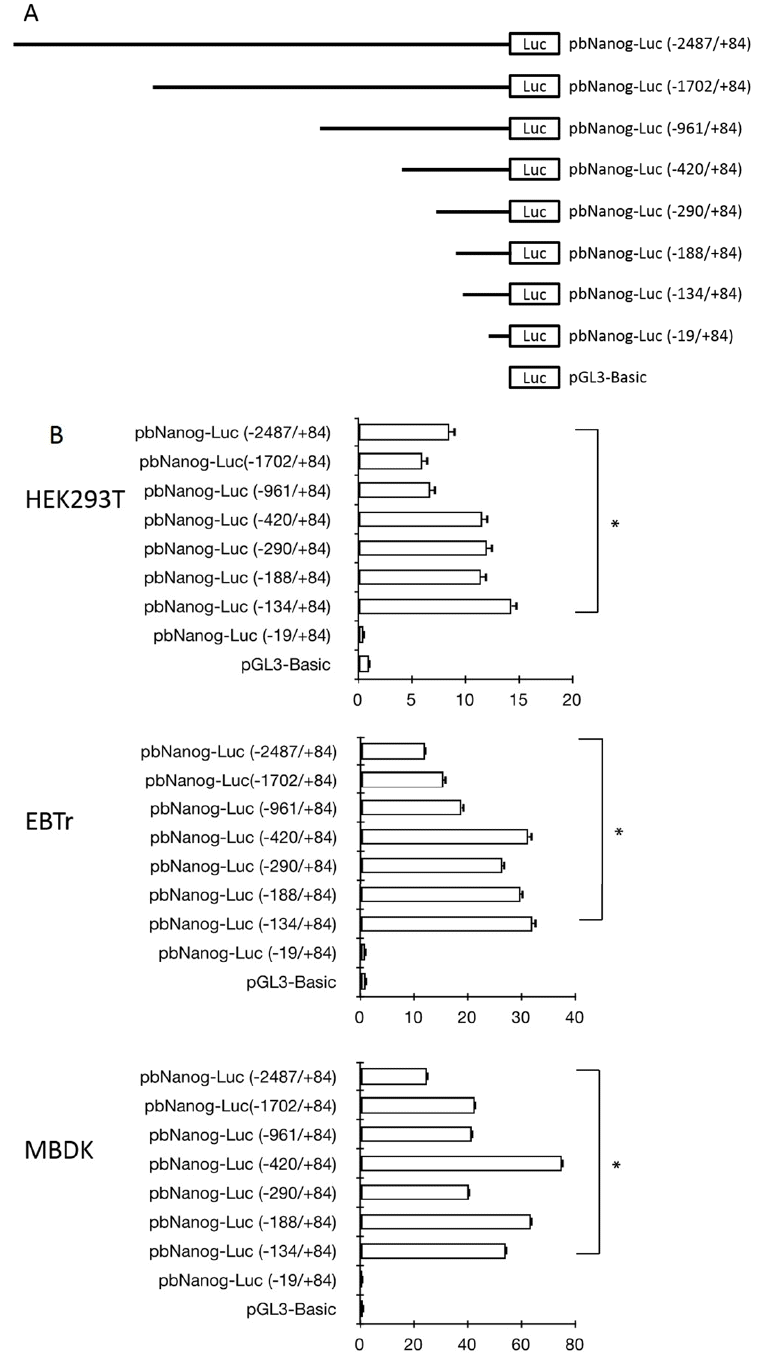
Sequence analysis of the 5′-proximal region of the bovine Nanog was performed using TRANASFAC and GENOMATIX software. Results showed the presence of DNA motifs recognized by known transcription factors in the 5′-proximal promoter region of the bovine Nanog gene (data not shown). It is notable that a 200-bp region immediately upstream of the transcription start site contained highly conserved transcription factor-binding motifs, including the Oct4/Sox2 binding site and Sp1 binding motifs. Results of sequence comparison between bovine and other mammalian Nanog promoters, e.g., human and murine, revealed another putative transcription factor binding site between Oct4/Sox2 and the transcription start site, e.g., one NF-κB transcription factor binding site and one Sp1 binding site between the NF-κB binding site and the transcription start site (Figure 2A). Of note, in murine, two Sp1 binding sites, Sp1A and Sp1B, which were identified in the Nanog proximal promoter region, were found to play a crucial role in transcription of the Nanog in murine ESC (Wu and Yao, 2006). Although the first Sp1 binding site was identified and was found to be similar to murine Sp1A, the second Sp1 binding region (Sp1B in murine) was not found in the bovine Nanog promoter (Figure 2A) suggesting differential transcriptional regulation of the Nanog in bovine. And also, question of whether this conserved NF-κB -binding site plays a role in Nanog transcription has not yet been investigated.
Effects of site-directed mutations within the NF-κB- and Sp1-binding region of the proximal promoter region of bovine Nanog. (A) Sequence alignment of bovine, human, and mouse Nanog 5′-proximal promoter sequences. Oct4/Sox2–binding site was marked in italic, NF-κB–bindsing site is in bold and Sp1A-binding site is in bold/italic. Sp1A was identified in this study, and was also identified by Wu and Yao (2006); Sp1B was identified by Wu and Yao (2006). (B) Role of NF-κB- and Sp1-binding sites in bovine Nanog promoter activity in HEK293T cells. Wild-type or mutants of the bovine Nanog 5′-deletion construct (pbNanog-Luc [−134/+84], pbNanog-Luc [−134/+84]-mutSp1, and pbNanog-Luc [−134/+84]-mutNF-κB) (left) were co-transfected with pRL-Tk into HEK293T cells and cultured for two days. Each activity value represents the average of at least three independent experiments and is normalized according to the activity of co-transfected Renilla (* p<0.05). (C) Comparison of Nanog 5′-flanking activity between and wild- and Sp1 mutant type of GFP reporter vector. HEK293T cells, human embryonic kidney cell line.
To investigate whether putative NF-κB- and Sp1 binding sites regulate bovine Nanog promoter activity, we introduced mutations to the NF-κB (GAATTC → TCTAGA) and Sp1 (GGCGGG → TCTAGA) binding sites to generate the pbNanog-Luc (−134/+84)-mutNF-κB and pbNanog-Luc (−134/+84)-mutSp1 constructs, respectively (Figure 2B). Ablation of the Sp1 binding site resulted in diminished Nanog promoter activity by approximately 50% (p<0.05), whereas mutation of the NF-κB binding site had no impact on reporter activity (Figure 2B). These data suggest a role of the Sp1 binding site in bovine Nanog expression; in addition, the NF-κB binding site might be dispensable for transcriptional activity of the Nanog promoter (Figure 2B). In addition, these findings were also confirmed in a transfection experiment with murine ESCs, where a GFP construct under the bovine Nanog promoter carrying Sp1 mutation was transfected and resulted diminished GFP expression, whereas GFP expression with wild type bovine Nanog promoter was preserved (Figure 2C).
Direct binding of Sp1 to the Sp1-binding site in the bovine Nanog promoter
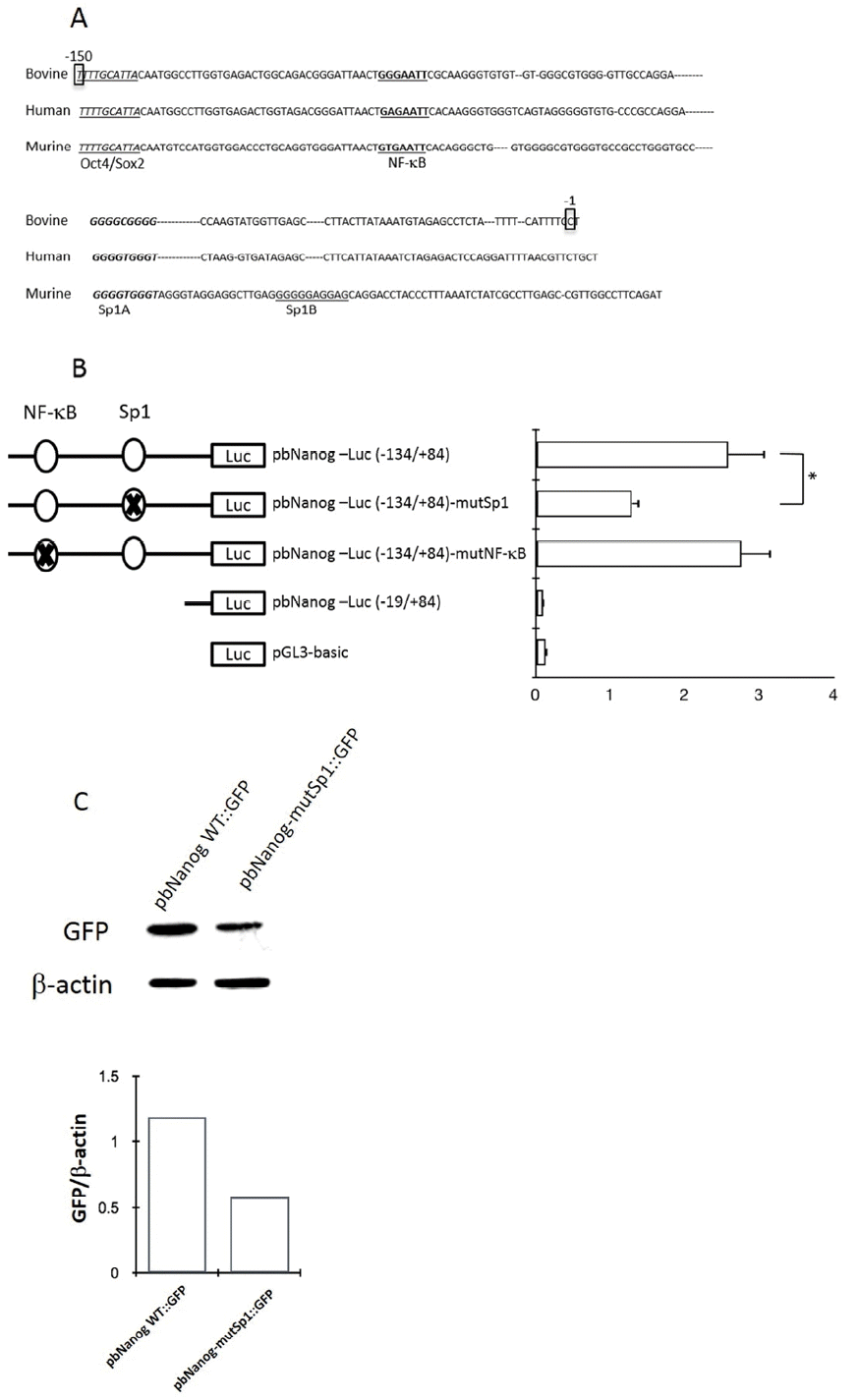
To evaluate the ability of Sp1 to recognize the Sp1 binding site within the bovine Nanog promoter, EMSA was performed using a 26-bp probe spanning the Sp1-binding site combined with nuclear extracts from HEK293T cells and R1 ESCs. Use of this probe resulted in formation of a protein-DNA complex, which was found to compete in the presence of a 100-fold excess of the unlabeled wild-type probe (Figure 3A). The presence of the mutantSp1 probe (mutSp1) showed significantly diminished formation of the protein-DNA complex (Figure 3A). Supershift assay with Sp1 antibody further confirmed the Sp1-DNA complex formation (Figure 3B). Taken together, these data suggest the direct regulation of bovine Nanog proximal promoter activity by Sp1 transcription factor.
Gel shift analyses of nuclear protein binding to Sp1 probes. Results from incubation of Biotin 3′-end labeled Sp1 probes and mutated probes (Sp1-mt) with nuclear extract proteins from HEK293T cells. (A) Lane 1, Biotin labeled Sp1 probes incubated without HEK293T nuclear extract proteins; Lane 2, Sp1 probes incubated with nuclear extract; Lane 3, Sp1-mt probes incubated with nuclear extract; Lanes 4, specific competition with unlabeled Sp1-mt probes. (B) Supershift assay using Sp1 Ab. Lane 1, without nuclear extract; Lane 2, Sp1 probes with nuclear extract; Lanes 3 and 4, specific and nonspecific competition; Lane 5, Supershift assays using Sp1 Ab. Sp-1mut: Sp1 mutant probe; NS: nonspecific competitor; HEK293T NE, HEK293T cell nuclear extracts.
Physical occupation of Sp1 in the Sp1-binding site in the bovine Nanog proximal promoter by ChIP assay
To determine if Sp1 physically occupies the bovine Nanog proximal promoter, we performed ChIP assay with the nuclear fractions of MDBK cells. ChIP enrichment with Sp1 antibody which resulted in 212 bp amplicon across the bovine Nanog promoter showed Sp1 enrichment, whereas the control ChIP with a nonspecific antibody showed no enrichment (Figure 4).

Chromatin immunoprecipitation assay demonstrating the in vivo potential of Sp1 protein to bind to Nanog 5′-flanking region. Lane 1, input only; Lane 2, input with Sp1 antibody; Lane 3, input with isotype antibody; Lane 4, no template.
In this study, we isolated and analyzed transcription activity of the 5′-proximal region of the bovine Nanog promoter, resulting in identification of highly conserved cis-regulatory sequences, e.g. Oct4/Sox2-, NF-κB, and Sp1-binding sites within a 150-bp region upstream of the transcription start site. Using a promoter assay, we demonstrated markedly attenuated bovine Nanog promoter activity by site-directed mutation of the Sp1-binding site, suggesting that the Sp1-binding site is indispensable for bovine Nanog promoter activity in cells used for this study. In addition, results from the EMSA experiment demonstrated the ability of Sp1 to bind directly to the Sp1-binding site; this binding was diminished by use of either a competitor or mutant Sp1 probes. These data suggest that Sp1 can regulate bovine Nanog transcription by direct binding to the Sp1-binding site found within a 150-bp region of the proximal Nanog promoter. Furthermore, ChIP showed the physical occupation of Sp1 in the bovine Nanog proximal promoter containing Sp1 binding sites. These findings were further strengthened in transfection study, where GFP expression was derived by the bovine Nanog proximal promoter with a Sp1 mutation in murine ESCs (Figure 2C).
DISCUSSION
In murine ESCs, GFP expression driven by belonging to the Sp1-like/KLF family of transcription factors, is a ubiquitously expressed transcription factor, which regulates expression of a large number of genes having GC-rich promoters. Sp1-like/KLF transcription regulators may participate in diverse cellular functions, including self-renewal of ESC, cell proliferation, apoptosis, differentiation, and carcinogenesis (Black et al., 2001; Bouwman and Philipsen, 2002; Kaczynski et al., 2003). In particular, participation of Sp1 and Sp3 in self-renewal of ESC occurs by transcriptional regulation of the regulators for stemness, such as Oct4 and Nanog (Pesce et al., 1999; Wu and Yao, 2006). Results of sequence analysis also revealed the presence of a putative Sp1/KLF-binding site around 200 bp region upstream of the transcription start site (data not shown), suggesting that, like the human Nanog, this site might act as a binding site for members of the KLF family, such as KLF4 and Sp3 (Wu and Yao, 2006; Chan et al., 2009). Previous studies demonstrated that other KLF members, including KLF2, KLF4, and KLF5, are required for self-renewal of ESC in vitro and are functionally redundant in vitro and in vivo as well (Jiang et al., 2008; Chan et al., 2009). These transcription factors may also share many of the targets of Nanog, suggesting that these factors may share a close functional relationship (Jiang et al., 2008). In addition, KLF1 and KLF2, as well as KLF4 and Sp3, can activate promoter activity of human Nanog (Chan et al., 2009), supporting the functional redundancy of Sp1-like/KLF members for self-renewal of ESC. Our results are in agreement with those of previous studies in murine ESCs (Wu and Yao, 2005; 2006), suggesting that transcriptional regulation of Nanog expression by Sp1 might be evolutionarily conserved. However, it is worth noting that the proximal Nanog promoter for mouse contains two Sp1-binding sites, whereas the bovine Nanog promoter contains one Sp1-binding site. The functional consequence of the unique configuration of the bovine Nanog promoter, compared to murine or human Nanog promoter, is not certain.
Results of our study also showed that the NF-κB binding site, which is also evolutionarily conserved, is dispensable for bovine Nanog promoter activity in cells which were used in this study. Inhibition of canonical of NF-κB pathways was found to be associated with increased expression of Nanog and Oct4, and suppression of differentiation toward all primitive extraembryonic and embryonic lineages except primitive ectoderm and ectodermal lineages (Torres and Watt, 2008; Yang et al., 2010). This finding suggests that the NF-κB signaling pathway regulates stemness of mESC by suppressing differentiation of mESCs to somatic cells. However, direct binding of NF-κB transcription factor with the Nanog promoter and the significance of this binding has not been investigated. Therefore, further investigation is warranted in order to reveal the role of the NF-κB -binding site in Nanog expression in bovine ESC.
The gene regulatory network is considered to be functionally conserved for the purpose of establishing and maintaining the stemness of ESC and ICM in mammalian species. Results from analysis of transcription factor binding sites within the proximal region of the mammalian Nanog, which were identified in this study and other studies, revealed a high degree of conservation and common transcription binding sites, including Oct4/Sox2, NF-κB, and Sp1-binding sites, supporting the high degree of conservation of the gene regulatory network for Nanog transcription. It is worth noting that among them, the Sp1-binding site was found to be functionally preserved in the Nanog proximal promoter region from both mouse and bovine. However, despite the existence of the conserved genetic regulatory circuit for Nanog expression, the possible existence of a species specific regulatory circuit might explain the differential expression profiles of ICM among mammalian species and the differential requirements of cell culture conditions for each species, e.g., differential requirement of the LIF-STAT3 signaling pathway between mESC and hESC (Ginis et al., 2004). In this regard, it is reasonable to assume that the presence or absence of the downstream Sp1 site may aid in understanding of species-specific regulation of Nanog expression.
CONCLUSION
Our results demonstrate that the Sp1 transcription factor can regulate activation of the bovine Nanog promoter through direct binding to a Sp1-binding site within the proximal bovine promoter region. These findings extend the observations made in studies using mouse and human ESC. This study might provide a unique opportunity to gain an understanding of general transcriptional regulatory mechanisms that operate for self-renewal and suppression of mammalian ESC by members of the Sp1/KLF transcription factors.
ACKNOWLEDGMENTS
This research was supported in part by a grant from the Next-Generation 21 Program (Project No. PJ01109301) and Inha University Research Grant (INHA-50486).
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.