Isolation and Characterization of a New Methanobacterium formicicum KOR-1 from an Anaerobic Digester Using Pig Slurry
Article information
Abstract
A new methanogen was isolated from an anaerobic digester using pig slurry in South Korea. Only one strain, designated KOR-1, was characterized in detail. Cells of KOR-1 were straight or crooked rods, non-motile, 5 to 15 μm long and 0.7 μm wide. They stained Gram-positive and produced methane from H2+CO2 and formate. Strain KOR-1 grew optimally at 38°C. The optimum pH for growth was 7.0. The strain grew at 0.5% to 3.0% NaCl, with optimum growth at 2.5% NaCl. The G+C content of genomic DNA of strain KOR-1 was 41 mol%. The strain tolerated ampicillin, penicillin G, kanamycin and streptomycin but tetracycline inhibited cell growth. A large fragment of the 16S rRNA gene (~1,350 bp) was obtained from the isolate and sequenced. Comparison of 16S rRNA genes revealed that strain KOR–1 is related to Methanobacterium formicicum (98%, sequence similarity), Methanobacterium bryantii (95%) and Methanobacterium ivanovii (93%). Phylogenetic analysis of the deduced mcrA gene sequences confirmed the closest relative as based on mcrA gene sequence analysis was Methanobacterium formicicum strain (97% nucleic acid sequence identity). On the basis of physiological and phylogenetic characteristics, strain KOR-1 is proposed as a new strain within the genus Methanobacterium, Methanobacterium formicicum KOR-1.
INTRODUCTION
Methanogens are one of the key populations in methanogenic waste and wastewater treatment processes because they are responsible for the final step of the degradation of organic substances (Garcia et al., 2000). They are commonly isolated from natural anoxic environments, including freshwater and marine sediments, wet and waterlogged soils, the rumen and intestinal tract of animals and the gut of insects (Garcia, 1990; Morvan et al., 1996). In addition, molecular surveys targeting the 16S rRNA gene and mcrA gene, encoding the alpha subunit of methyl coenzyme M reductase, have revealed that numerous unidentified methanogens may also exist in such ecosystems (Sly et al., 1986; Chouari et al., 2005).
Biological methanogenesis occurs during the anaerobic treatment of sewage sludge, agricultural, municipal and industrial wastes, and manure of cattle, pigs and chickens. This anaerobic digestion process stabilizes the sludge with the formation of methane and offsets the costs of waste disposal. In some urban and industrial waste treatment plants the generated methane is used as an energy source. Anaerobic digestion technology in South Korea has been of greater interest after a ban on ocean dumping of animal manure, sewage sludge and food waste. There are about 49 biogas plants in South Korea that are generally recognized as economically and technically unsuccessful because the existing biogas plants are not running well, produce less methane than expected, or operation costs are very high (Kim et al., 2012). Therefore, there is a need to improve anaerobic digestion technologies, especially understanding microorganisms in anaerobic digesters using biomass such as pig slurry and food waste originally produced in South Korea.
Methanogens are difficult to study through culture-based methods, and therefore many researchers have instead used culture-independent techniques such as real-time polymerase chain reaction (qPCR), denaturing gradient gel electrophoresis (DGGE) and sequencing, which have proven to be valuable tools for studying the biodiversity of complex microbial communities such as those in anaerobic digesters (Nettmann et al., 2008; Cardinali-Rezende et al., 2012). To our knowledge, there is no published information available on a methanogen isolated from an anaerobic digester in South Korea. Hence, the present study was aimed to isolate and characterize methanogens responsible for methane production during anaerobic digestion of pig slurry.
MATERIAL AND METHODS
Source of inoculum
An anaerobic digestate sample (2 L) was collected in May 2011 from an anaerobic digester in Biogas Research Center, Hankyong National University, Anseong, Republic of Korea, which was built in early 2008 and kept running for about 4 years. The anaerobic digester had a volume of 150 m3 for anaerobic fermentation and produced 160 m3 biogas per day. The main fermentation substrate was pig slurry from a pig farm breeding 7,000 pigs. The characteristics of the anaerobic digestate in the digester showed the pH of 8.2, total volatile fatty acid concentration of 4,500 ppm and chemical oxygen demand of 70,000 ppm. The anaerobic digestate of this digester was used as inoculum for isolation of methanogen.
Medium
Methanogens are extremely sensitive to oxygen and require strict anoxic condition and pre-reduced media are essential for their growth and isolation. The methods used for the preparation of media and substrate solutions and the cultured techniques were those of Hungate (1950), as modified by Balch et al. (1979). Basal medium for enrichment culture and isolation was prepared with modification as described by Sowers and Schreier (1995) and contained the following compounds (per liter distilled water): KH2PO4, 0.5 g; MgSO4·7H2O, 0.4 g; NaCl, 0.4 g; NH4Cl, 0.4 g; CaCl2·2H2O, 0.05 g; FeSO4·7H2O, 0.002 g; trace element solution, 1.0 mL; vitamin solution, 10.0 mL; yeast extract, 1.0 g; sodium acetate, 1.0 g; sodium formate, 2.0 g; sludge fluid, 50.0 mL; NaHCO3, 4.0 g; fatty acid mixture, 20.0 mL; resazurin, 0.001 g; cysteine hydrochloride, 0.5 g; and Na2S·9H2O, 0.5 g. The fatty acid mixture contained valeric acid, 0.5 mL; isovaleric acid, 0.5 mL; 2-methylbutyric acid, 0.5 mL; isobutyric acid, 0.5 mL; and distilled water, 20.0 mL, adjusting pH to 7.5 with concentrated NaOH. The trace element solution was made up of (in one liter of distilled water): nitrilotriacetic acid, 1.5 g; MgSO4·H2O, 3.0 g; MnSO4·H2O, 0.5 g; NaCl, 1.0 g; FeSO·7H2O, 0.1 g; CoCl2·6H2O, 0.1 g; CaCl2, 0.1 g; ZnSO4·7H2O, 0.1 g; CuSO4·5H2O, 0.01 g; Al2(SO4)3·12H2O, 0.01 g; H3BO3, 0.01 g; Na2MoO4·2H2O, 0.01 g; NiSO4·6H2O, 0.03 g; Na2SeO3, 0.02 g; and Na2WO4·2H2O, 0.02 g. Firstly, 1.5 g nitrilotriacetic acid was added to 500 ml distilled water, adjusting pH to 6.5 with KOH and then minerals were added and dissolved making the solution up to a volume of 1,000 mL. The vitamin solution was made up of (in one liter of distilled water): folic acid, 0.01 g; riboflavin, 0.03 g; L-6,8-thioctic acid, 0.03 g; cyanocobalamin, 0.05 g; thiamine chloride hydrochloride, 0.1 g; pyridoxine hydrochloride, 0.15 g; hemicalcium D-(+)-pantothenate, 0.05 g; nicotinic acid, 0.1 g; D-(+)-biotin, 0.01 g; and 4-aminobenzoate, 0.04 g. Clarified sludge fluid was prepared by filtering the anaerobic digestate collected from the anaerobic digester, centrifuging at 13,000×g for 30 min, autoclaving at 121°C for 20 min and then recentrifuging to obtain a clear yellow solution. The sludge fluid (5%, v/v) was added as a source of unknown growth factors. The medium was prepared under a H2/CO2 (80:20, v/v) gas phase at 173 kPa (25 psi). The medium was sterilized by autoclaving at 121°C for 30 min.
Enrichment and isolation
Enrichments were performed in the basal medium adjusted pH 7.0 under the H2/CO2 (80:20, v/v) gas phase. Vials containing 45 mL medium were inoculated with 5 mL inoculum. Benzyl penicillin (2×1010 IU/L) and streptomycin sulphate (7×104 IU/L) were added to inhibit the growth of non-methanogenic organisms. The inoculated culture vials were incubated at 38°C in the dark for two weeks. A high amount of methane was detected in the culture, and then 5 mL of the culture was anaerobically transferred into a new vial of sterile basal medium. After four successive transfers, roll tubes containing the basal medium with 1.8% agar were prepared. Well-isolated colonies were withdrawn with Pasteur pipettes and transferred to culture tubes containing basal medium under anaerobic condition. The culture tubes were sealed with butyl-rubber stoppers and repressurized with sterile filtered H2/CO2 (80:20, v/v) at 173 kPa (25 psi). The organism was re-isolated with the solid basal medium from the liquid cultures. To check the purity of methanogen culture, it was inoculated on bacterial growth medium. The bacterial growth medium was prepared by adding peptone (2.5 g/L), yeast extract (2.5 g/L), D-glucose (0.5 g/L), D-cellobiose (0.5 g/L), D-xylose (0.5 g/L) to the basal medium. For the further purification of isolated strain KOR-1, a mixture of four antibiotics was used (benzyl penicillin, 0.5 mg/mL; streptomycin sulphate, 0.5 mg/mL; vancomycine-HCl, 0.2 mg/mL; and ampicillin, 0.2 mg/mL). After purification, the isolate was kept in the basal medium without antibiotics. We isolated three methanogens from the isolation process. Strain KOR-1 was microscopically distinguished with rod shape compared with coccal shape of other isolated methanogens. Thus characteristics of the isolated strain KOR-1 were studied at the first.
Physiological studies
Substrate utilization, growth temperature range, NaCl concentration range, pH range, antibiotics tolerance and DNA G+C content were investigated for the physiological characteristics of strain KOR-1. Growth was determined by observing optical density (OD) at 660 nm (OD660) with a spectrophotometer (V-530, Jasco, Tokyo, Japan) and measuring the concentration of methane in the gas phase with a gas chromatograph (GC-2010A, Shimadzu, Kyoto, Japan). All experiments for the physiological studies were repeated twice.
The basal medium with acetate and formate omitted, was prepared under N2, and used for substrate-utilization studies. Anaerobic stocks of filter-sterilized substrates (sodium formate, sodium acetate, trimethylamine, methanol, 2-propanol, and isobutanol) were prepared and added separately at final concentrations of 50 mM. A freshly grown culture was inoculated at 10% (v/v) and vials were incubated at 38°C for 20 days. Medium under H2/CO2 served as the control.
The optimal growth temperature was determined in the basal medium at the optimal pH. Vials inoculated with 10% (v/v) culture were incubated at temperatures in the range from 20°C to 50°C. The vials were pressurized every other day with H2/CO2 to ensure an adequate supply of substrate.
The pH requirement was determined at the optimum temperature in the basal medium adjusted to pH values ranging from 4.0 to 9.0. Values above pH 4.0 were produced by adding sterile Na2CO3 to medium of pH 4.0 until the required value was reached. The pH 4.0 medium was prepared by omitting NaHCO3 from basal medium and cooling it under a CO2 headspace.
Sensitivity of strain KOR-1 to ampicillin, penicillin G, spectromycin, kanamycin, tetracycline and chloramphenicol (all with concentration of 100 μg/mL) was tested. Five mL aliquots of the cultures were transferred into fresh medium containing one of the four antibiotics. Strain KOR-1 was incubated for 1 week at 38°C. The impact of antibiotics was determined by comparing the growth of cultures containing these antibiotics with control.
The salinity range of the isolate was tested at NaCl concentrations from 0.5% to 3.0% NaCl at 0.5% intervals. Media with various concentrations of NaCl were prepared by adding sterile anoxic stock solution of 58.44 g/L NaCl to medium.
Microscopy
An Olympus BX41 phase-contrast microscope (Olympus, Tokyo, Japan) was used routinely to observe cells. The Gram character was determined using a standard Gram-stain kit (BBL Microbiology Systems, Becton Dickinson, Bergen County, NJ, USA). Motility was determined by the hanging-drop method, using a glass cavity slide.
DNA extraction and G+C content
Culture samples of strain KOR-1 grown with the basal medium were used for DNA isolation (FastDNA SPIN kit for soil, MP Biomedicals, Irvine, CA, USA) and following the manufacturer’s instructions. DNA integrity was checked on 1% agarose gel and DNA concentration was determined using Nanodrop (ND 2000, Thermo Fisher Scientific, Waltham, MA, USA). The DNA G+C content was determined from thermal denaturation profiles (Sly et al., 1986). The experiments for the DNA extraction and G+C content of strain KOR-1 were conducted several times until to get clear results.
PCR amplification of 16S rRNA genes
16S rRNA gene amplicons (~1,350 bp) were obtained using the following primer pair: Ar109f (5′ ACKGCTCAGTAACACGT-3′) (Großkopf et al., 1998) and Ar1383r (5″-CGGTGTGTGCAAGGAGCA-3′) (Shlimon et al., 2004). Reaction mixtures contained the following components in a final volume of 20 μL in a 200 μL PCR reaction tube: 2 μL PCR reaction buffer (Takara, Tokyo, Japan), 2 μL dNTP mix (35 mM), 0.5 μL each primer (10 pmol/μL), 0.1 μL Taq DNA polymerase (Takara, Japan), 1.0 μL template DNA sample (100 ng) and 13.9 μL molecular grade water (Severn Biotech Ltd, Kidderminster, Worcester, UK). The PCRs were started by immediately placing the reaction tubes into the preheated (94°C) thermal cycler (PCR Thermal Cycler Dice, Takara, Japan). The thermal program was as follows: an initial denaturation step (94°C, 4 min) was followed by 30 cycles of denaturation (94°C, 30 s), annealing (55°C, 30 s) and extension (72°C, 90 s). After a final extension step (72°C, 6 min) samples were kept at 4°C until further analysis. Each PCR run included a positive control using DNA extracted from pure cultures and a negative PCR control where molecular grade water was substituted for the DNA template. The DNA product was analyzed using gel electrophoresis (1.2% w/v agarose gel stained and run at 100 V for 30 min in 1×Tris-acetate-ethylene-diamine-tetracetic acid (TAE) buffer with 5 μL aliquots of each DNA product. The TAE buffer was composed of 40 mM Tirs base, 20 mM acetic acid and 0.5 M ethylene-diamine-tetracetic acid (EDTA) (pH 8.0). The gel was run with 2.5 μL Ladder I DNA quantification marker (TNTresearch, Seoul, Korea). The gel was imaged using Bio Imaging System (Daihan, Seoul, Korea) and photograph was taken using WiseCapture II software (Daihan, Korea).
PCR amplification of mcrA genes
The mcrA genes fragments were amplified using the primer combinations MLf (5′-GGTGGTGTMGGATTC ACACARTAYGCWACAGC-3′) and MLr (5′-TTCA TTGCRTAGTTWGGRTAGTT-3′), yielding ~490 bp amplicons (Luton et al., 2002), ME1 (5′-GCMAT GCARATHGGWATGTC-3′) and ME2 (5′-TCATKGCRTA GTTDGGRTAGT-3′), yielding ~740 bp amplicons (Hales et al., 1996), and MR1 (5′-GACCTCCACTWCGT VAACAACGC-3′) and ME2, yielding amplicons of ~1,100 bp (Simankova et al., 2003). Denaturation, annealing and extension were carried out at 96°C (15 s), 55°C (30 s) and 72°C (90 s), respectively, with MLf/MLr primers, and 94°C (40 s), 50°C (45 s) and 72°C (90 s), respectively, with ME1/ME2 primers and MR1/ME2 primers.
Phylogenetic and sequencing analysis
PCR products were purified with the AccuPrep PCR purification kit (Bioneer, Daejeon, Korea). Sequencing of PCR products was performed using the BigDye terminator cycle sequencing kit on ABI 3730XL capillary DNA Sequencer (Applied Biosystems, Thermo Fisher Scientific Inc., Carlsbad, CA, USA). 16S rRNA and mcrA genes sequences of strain KOR-1 were compared with the other similar sequences obtained from GenBank using the BLAST program. Phylogenetic analysis was conducted in MEGA 4.0 (Tamura et al., 2007). Eight additional 16S rRNA sequences (Methanobacterium formicicum [NR025028]; Methanobacterium subterraneum [NR028247]; Methanobacterium palustre [NR041713]; Methanobacterium alcaliphilum [NR028228]; Methanobacterium bryantii [NR042781]; Methanobacterium oryzae [NR028171]; Methanobacterium ivanovii [NR041716]; Methanosphaera stadtmanae [NR028236]) representing methanogens were included in phylogenetic analysis. Phylogeny was further confirmed by mcrA gene and mcrA protein sequences (Methanobacterium formicicum [EF465103]; Methanobacterium palustre [AB542760]; Methanobacterium kanagiense [AB551870]; Methanobacterium bryantii [AF313806]; Methanobacterium aarhusense [AY386125]; Methanopyrus kandleri [U57340]).
Nucleotide sequence accession number
The 16S rRNA, mcrA gene and mcrA amino acid sequences of strain KOR-1 determined in this study have been deposited in the GenBank database under No JQ973735, JX141395, and AFP23434.
RESULT AND DISCUSSION
Characterization of the isolated methanogen
A new methanogen was isolated from an anaerobic digester using pig slurry from Anseong, South of Korea. The methanogenic enrichment culture dominated by long rods was obtained after two months. Visible colonies in agar tubes appeared after three weeks of incubation at 38°C. Surface colonies were about 2 mm in diameter, yellow, circular and concave after three weeks incubation. Only one strain, designated KOR-1, was characterized in detail.
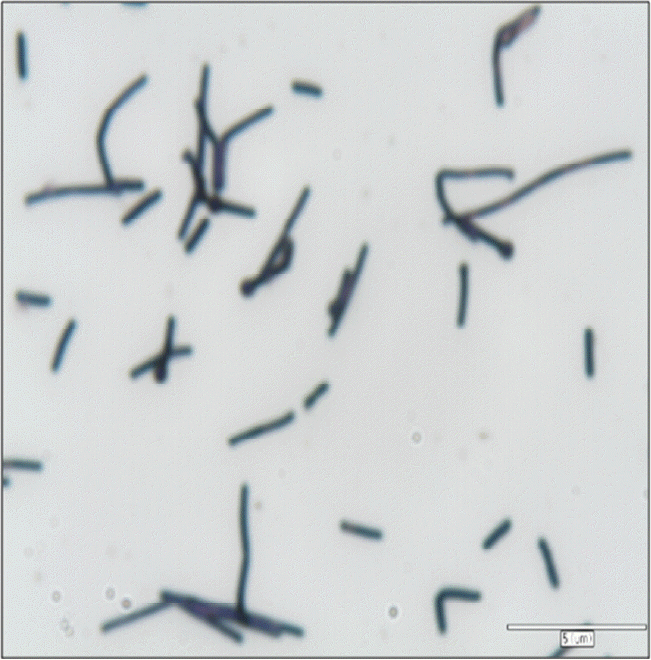
Table 1 shows phenotype and growth characteristics of strain KOR-1 comparing with Methanobacterium formicicum DSM1535T (Bryant and Boone, 1987) and Methanobacterium bryantii DSM 863T (Bryant et al., 1967; Boone, 1987). The isolate was phenotypically similar to the compared species of Methanobacterium. Cells of KOR-1 were straight or crooked rods, non-motile, 5.0 to 15.0 μm long and 0.7 μm wide. They stained Gram-positive (Figure 1). In liquid cultures, cells of KOR-1 often attached to particles, forming clumps. Along with Methanobacterium formicicum DSM1535T and Methanobacterium bryantii DSM 863T, strain KOR-1 demonstrated wide growth temperature and pH ranges. Growth of strain KOR-1 was observed in the temperature range of 25°C to 50°C, with fastest growth at 38°C. Most strains in the genus Methanobacterium are mesophilic and grow in a wide range of temperature from 3°C to 50°C (Krivushin et al., 2010) but not at 55°C (Joulian et al., 2000; Boone, 2001), distinguishing them from the related thermophilic genus, Methanothermobacter (Wasserfallen et al., 2000). The optimum pH for growth was 7.0 for strain KOR-1. No growth occurred below pH 6.0 or above 8.3. The optimum pH for growth of strain KOR-1 (pH 7.0) is similar to those of Methanobacterium formicicum (pH 6.6 to 7.8) (Bryant and Boone, 1987) and Methanobacterium congolense (pH 7.2) (Cuzin et al., 2001) and slight different from those of Methanobacterium subterraneum (pH 7.8 to 8.8) (Kotelnikova et al., 1998), Methanobacterium alcaliphilum (pH 8.1 to 9.1) (Worakit et al., 1986) and Methanobacterium espanolae (pH 5.6 to 6.2) (Patel et al., 1990). The growth range for pH of strain KOR-1 falls within the range that has been reported for the genus Methanobacterium (Krivushin et al., 2010). Strain KOR-1 grew at NaCl concentrations of 0.5% to 3.0%, with optimum growth at 2.5% NaCl. The range is typical for a halotolerant organism.

Comparative characteristics of strain KOR-1, Methanobacterium formicicum DSM1535T and Methanobacterium bryantii DSM 863T
Strain KOR-1 used H2/CO2 and sodium formate (50 mM) to produce methane. Under a N2:CO2 (4:1) atmosphere, strain KOR-1 could not produce methane from sodium acetate (50 mM), trimethylamine (50 mM), methanol (50 mM), 2-propanol (50 mM) or isobutanol (50 mM). The substrate utilization for methanogenesis of strain KOR-1 is similar to that of Methanobacterium formicicum DSM1535T (Bryant and Boon, 1987), while Methanobacterium bryantii DSM863T utilized 2-propanol and isobutanol as hydrogen donors for the reduction of CO to produce methane in addition to H2/CO2 (Bryant et al., 1967; Boone, 1987). Strain KOR-1 shared phenotypic characteristics of the strain DSM1535 of Methanobacterium formicicum isolated from digester sludge but had a growth requirement for sludge fluid or yeast extract, a characteristic not found in the type strain. Strain KOR-1 is also similar to Methaonbacterium oryzae DSM 11106T (Joulian et al., 2000) and Methanobacterium beijingense DSM 15999T (Ma et al., 2005), both of which can use H2/CO2 and formate for growth, requiring yeast extract. Thus, the growth of strain KOR-1 is common among members of the genus Methanobacterium, considering the strain as chemoautotrophic (Boone, 2001).
The strain tolerated ampicillin, penicillin G, kanamycin and streptomycin and was sensitive to chloramphenicol. However, tetracycline completely inhibited cell growth. Tetracycline is related to inhibition of translation during protein biosynthesis. Hilpert et al. (1981) reported that tetracycline inhibited growth of Methanococcus vannielii, but most methanogens tested were insensitive. Chopra and Howe (1978) reported that methanogens used in their study were insensitive to tetracycline because the growth medium used contained high magnesium. They indicated that the tetracycline susceptibility of certain Gram-negative bacteria was decreased as the concentration of magnesium in a medium is increased, probably through the formation of antibiotic-cation complexes. The basal medium for the growth of strain KOR-1 containing 3.0 mg MgSO4/L medium may not be enough to decrease its susceptibility to tetracycline. Whitman et al. (2014) reported that members of the genus Methanobacterium may possess a particulate dehydrogenase enzyme that is sensitive to chloramphenicol.
The G+C content of genomic DNA of strain KOR-1 was 41 mol%. This value is similar to Methanobacterium formicicum DSM1535T (Bryant and Boone, 1987), but higher than Methanobacterium bryantii DSM863T (Bryant et al., 1967; Boone, 1987). The family Methanobacteriaceae has a wide range of the mol% G+C of the DNA between 23 and 62 (Oren, 2014).
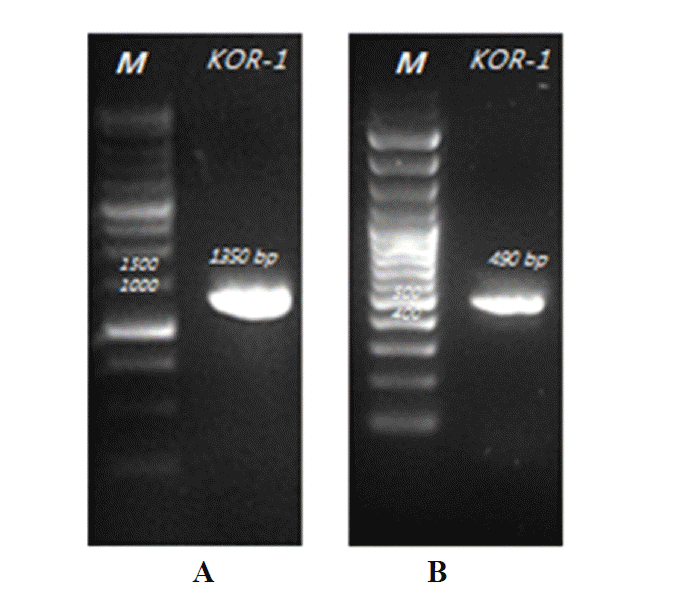
Molecular characterization
PCR based on 16S rRNA gave an amplicon size of 1,350 bp (Figure 2a). The identification was also confirmed by mcrA gene-based amplification using MLf and MLr primers that resulted in product size of 440 to 490 bp (Figure 2a). A large fragment of the 16S rRNA gene (~1,350 bp) was obtained from the isolate and sequenced. Comparative 16S rRNA gene sequence analysis showed that strain KOR-1 was affiliated with the order Methanobacteriales. The closest relatives of strain KOR-1 were Methanobacterium formicicum (98%, sequence similarity), Methanobacterium bryantii (95%) and Methanobacterium ivanovii (Figure 3). For PCR amplification of the mcrA gene, we used primers ME1/ME2 (Hales et al., 1996) and MR1/ME2 (Simankova et al., 2003) to obtain the nearly full-length mcrA gene. The mcrA gene sequence also indicated that strain KOR-1 was a member of the order Methanobacteriales. The closest relatives on the bases on the mcrA gene sequence were Methanobacterium formicicum (97%) and Methanobacterium palustre (94%). In addition, we determined the sequence part of mcrA gene (440 to 490 bp) and then constructed a molecular phylogenetic tree based on methyl-coenzyme M reductase II alpha subunit sequences of mcrA gene. The mcrA gene sequence-based tree also indicated that strain KOR-1 was a member of the order Methanobacteriales. The closest relative as based on mcrA gene sequence analysis was Methanobacterium formicicum strain (97% nucleic acid sequence identity) (Figure 4). No major differences were observed between DNA and amino acid sequences or between the different algorithms used (Figures 3 to 5). The phylogenetic tree with 16S rRNA gene (Figure 3) sequences showed two major clusters: cluster 1 and 2, which were entirely different from each other. Cluster 1 contained 2 sub clusters: with KOR-1 in sub cluster 1 that showed 100% similarity with Methanobacterium formicicum (NR025028). mcrA gene (Figure 4) and amino acid sequences (Figure 5) showed three major clusters: cluster 1 contained 3 sub clusters with KOR-1 in sub cluster 1 that showed 99% similarity with Methanobacterium formicicum. All three showed similar phylogenetic results and thus are in accordance with Luton et al. (2002) who stated that mcrA gene sequence can be used an alternative to 16S rRNA based sequences.
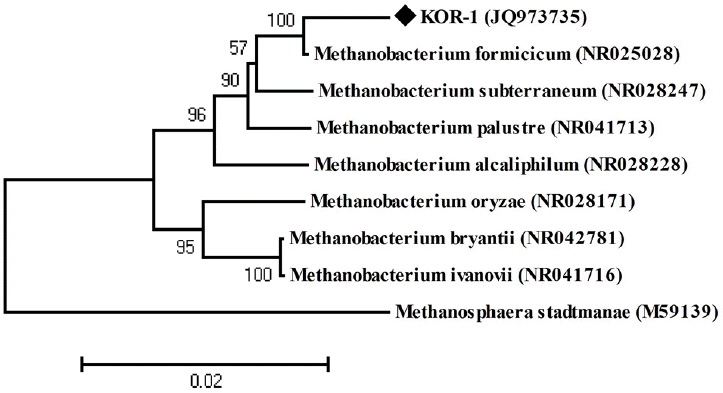
Phylogenetic dendrogram of 16S rRNA gene sequences showing the position of Methanobacterium formicicum strain KOR-1 relative to other species of the genus Methanobacterium as well as selected reference sequences of methanogens. Methanosphaera stadtmanae was used as outgroup references. The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al., 2007). Bootstrap values are shown at nodes (percentages of 500 replicates). GenBank accession numbers are indicated. A bar represents 0.02 substitutions per nucleotide position.
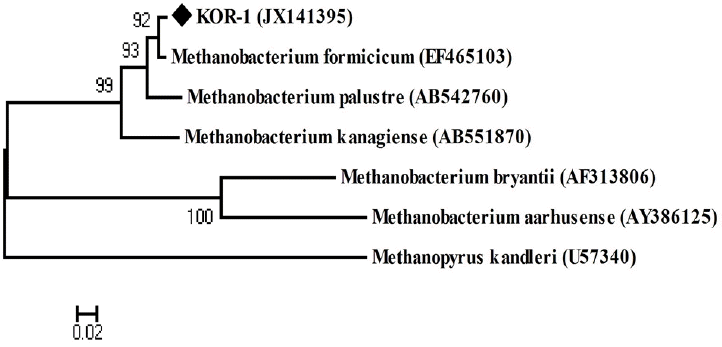
Phylogenetic tree of deduced mcrA genes sequences indicating the relationship of Methanobacterium formicicum strain KOR-1 to members of the genus Methanobacterium and other methanogenic archaea. Methanopyrus kandleri was used as outgroup references. GenBank accession numbers are indicated. Bootstrap values are shown at nodes (percentages of 500 replicates). A bar represents 0.02 substitutions per nucleotide position.
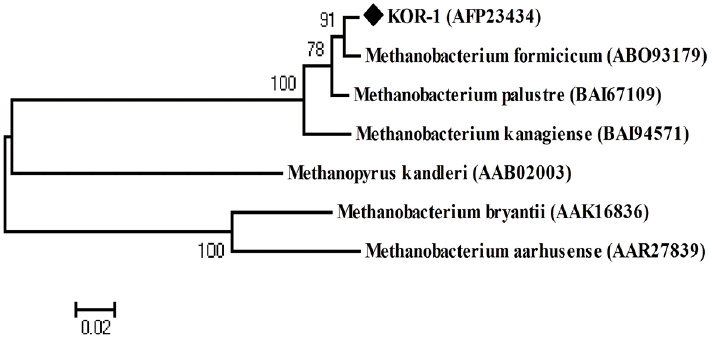
Phylogenetic tree of deduced mcrA amino acid sequences showing the relationship of Methanobacterium formicicum KOR-1 with related methanogenic archaea. The tree was constructed based on distance matrices (154 amino acid positions; Poisson correction) by the neighbor-joining method. The sequence of Methanopyrus kandleri was used as the outgroup reference. Accession numbers are shown in parentheses. A bar represents 0.02 substitutions per nucleotide position.
On the basis of physiological and phylogenetic differences, strain KOR-1 was proposed as a new strain within the genus Methanobacterium, Methanobacterium formicicum KOR-1.
ACKNOWLEDGMENTS
This work was supported by a research grant from Hankyong National University for an academic exchange program in 2014.
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.