Direct Interaction between Ras Homolog Enriched in Brain and FK506 Binding Protein 38 in Cashmere Goat Fetal Fibroblast Cells
Article information
Abstract
Ras homolog enriched in brain (Rheb) and FK506 binding protein 38 (FKBP38) are two important regulatory proteins in the mammalian target of rapamycin (mTOR) pathway. There are contradictory data on the interaction between Rheb and FKBP38 in human cells, but this association has not been examined in cashmere goat cells. To investigate the interaction between Rheb and FKBP38, we overexpressed goat Rheb and FKBP38 in goat fetal fibroblasts, extracted whole proteins, and performed coimmunoprecipitation to detect them by western blot. We found Rheb binds directly to FKBP38. Then, we constructed bait vectors (pGBKT7-Rheb/FKBP38) and prey vectors (pGADT7-Rheb/FKBP38), and examined their interaction by yeast two-hybrid assay. Their direct interaction was observed, regardless of which plasmid served as the prey or bait vector. These results indicate that the 2 proteins interact directly in vivo. Novel evidence is presented on the mTOR signal pathway in Cashmere goat cells.
INTRODUCTION
The mammalian target of rapamycin (mTOR) signaling pathway responds to nutrients, growth factors, the bioenergetic status of the cell, and cellular stress to coordinate the regulation of cell growth, metabolism, and differentiation. Ras homolog enriched in brain (Rheb) and FKBP38 are two important regulatory proteins in the mTOR pathway. Rheb was identified as an immediate early gene that encodes 184 amino acids with a deduced molecular mass of 20,497 Da in the hippocampus (Yamagata et al., 1994). Drosophila and yeast each have a single Rheb gene, whereas mammals have 2-termed Rheb1 and Rheb2/Ras homolog enriched in brain like 1 (Patel et al., 2003).
Rheb is a Ras-like small guanosine triphosphatase (GTPase) that shuttles between an active guanosine triphosphate (GTP)-bound form and an inactive guanosine driphosphate (GDP)-bound form. Rheb GTP loading and function are regulated through its GTP-/GDP-bound status (Lee et al., 2013). Rheb-GTP can activate mTOR through direct interaction with it (Duran et al., 2012; Yadav et al., 2013). Rheb1 predominates over Rheb2 in the regulation of mTORC1 in vivo (Zou et al., 2011; Bonneau et al., 2012). Rheb is essential for murine development (Goorden et al., 2011) and postnatal brain development (Zou et al., 2011); it also affects central brain neuronal morphology (Brown et al., 2012) and pigmentation (Zitserman et al., 2012) in Drosophila. Rheb signaling regulates mTORC1-dependent apoptosis (Karassek et al., 2012) and autophagy (Zhou et al., 2009; Sciarretta et al., 2012). These studies have demonstrated that Rheb regulates many cellular functions via mTOR signaling.
FK506 binding protein 38 (FKBP38) (also known as FKBP8), a member of the FKBP family, was isolated from a cDNA library of human T cells in the early 1990s (Lam et al., 1995). FKBPs constitute a family of proteins that catalyze the cis/trans interconversion of the peptide bond that precedes the proline residue. FKBP38 is a unique member of the FKBP family, in that it is membrane-anchored, and has peptidyl-prolyl cis-trans isomerase (PPIase) activity. FKBP38 lies in the membranes of mitochondria and the endoplasmic reticulum (ER) and mediates the biogenesis of membrane proteins on the cytoplasmic side of the ER membrane (Banasavadi-Siddegowda et al., 2011).
FKBP38 mediates neuronal apoptosis (Maestre-Martinez et al., 2011), and FKBP38−/− mice die soon after birth (Shirane et al., 2008). The catalytic domain of FKBP38 binds to Bcl-2 through a charge-sensitive loop (Haupt et al., 2012) to control apoptosis; this interaction is regulated by Ca2+ and Rheb (Ma et al., 2010; Maestre-Martinez et al., 2011; Haupt et al., 2012). FKBP38 is associated with many functions, from programmed cell death to the development of the embryo and postnatal brain.
FKBP38 was identified as an endogenous inhibitor of mTORC1 in vitro and in vivo; the mTORC1-FKBP38 association is enhanced in cells that have been deprived of amino acids or serum. Rheb-GTP binds to FKBP38 when amino acids are provided and prevents the mTORC1-FKBP38 interaction, affecting the release and activation of mTOR (Bai et al., 2007). The switch I region of Rheb is critical for its interaction with FKBP38 (Ma et al., 2008).
The activation of mTORC1 is impaired by FKBP38 and the FKBP38-Rheb interactions have been confirmed (Bai et al., 2007; Dunlop et al., 2009). Otherwise, overexpression or knockdown (Maehama et al., 2008; Wang et al., 2008) of FKBP38 does not affect the phosphorylation of mTORC1 substrates. Subsequent studies reported that FKBP38 is not involved in the Rheb-dependent activation of mTORC1 in vitro (Sato et al., 2009) and that the interaction between Rheb and FKBP38 could not be detected by 3 separate in vitro assays (Uhlenbrock et al., 2009).
Due to the discrepancies with regard to the interaction between Rheb and FKBP38, we cloned cashmere goat Rheb (GenBank accession HM569224) and FKBP38 (GenBank accession JF714970) genes and studied this relationship in cashmere goat cells by coimmunoprecipitation (co-IP) and yeast two-hybrid assay to resolve this controversy and examine the mTOR signaling pathway in cashmere goat cells.
MATERIALS AND METHODS
Cell culture conditions
Inner Mongolia cashmere goat fetal fibroblasts (GFb cells) were maintained as monolayer cultures in DMEM/F12 (Gibco, Paisley, PA49RF, Scotland, UK), supplemented with 10% fetal bovine serum (FBS), 100 U penicillin G/mL, and 100 mg streptomycin/mL (FBS, Hyclone Laboratories, Inc. Logan, UT, USA and penicillin/streptomycin, Sigma-Aldrich, Inc. St. Louis, USA). Cell cultures were maintained and incubated at 37°C in humidified air with 5% CO2.
Coimmunoprecipitation identification of protein interaction
We cloned the complete ORF of cashmere goat FKBP38 and Rheb and generated 2 expression vectors: pIRES2-DsRed2-FKBP38 and pIRES2-EGFP-Rheb. Goat fetal fibroblasts were cotransfected with pIRES2-DsRed2-FKBP38 and pIRES2-EGFP-Rheb with Lipofectamine 2000 and screened using G418. Control cells (non-transfected) and cotransfected cells were homogenized in M-PER Mammalian Protein Extraction Reagent (Pierce Coimmunoprecipitation Kit, Thermo, Rockford, USA), incubated at room temperature for 5 min, and spun at 13,000×g for 10 min at 4°C. The supernatants were isolated as total protein.
For the co-IP assay, the total proteins were transferred to Millipore catch and release spin columns (500 μg), which combined 3 μg anti-Rheb antibody (Santa Cruz Biotechnology Inc, Heidelberg, Germany), 10 μL affinity ligand, and Sufficient 1× wash buffer to a final volume of 500 μL. The column was incubated on a shaker at room temperature for 45 minutes. The eluates (including interacting proteins) were collected by centrifugation, washed 3 times with 1× denaturing elution buffer, and examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For the western blot, the proteins were separated by SDS-PAGE and transferred to Immun-Blot polyvinylidene fluoride (Bio-Rad, Hercules, CA, USA) membrane. The membrane was treated with 5% nonfat dry milk in Tris-buffered saline and 0.2% Tween 20 (TBST) at 37°C for 1 h and incubated overnight with primary antibody at 4°C in TBST and 2% nonfat dry milk. The membrane was washed 3 times with TBST and incubated with secondary antibody in TBST and 5% milk for 1 h at room temperature. After the membranes were washed 3 times with TBST (10 min each time), the membranes were developed with an enhanced chemiluminescence detection system (GE Healthcare, Buckinghamshire, UK).
Yeast two-hybrid analysis of protein interaction
A yeast two-hybrid screen was performed using Matchmaker galactose-regulated upstream promoter element 4 (GAL4) Two-Hybrid System 2 (Clontech, BD Biosciences, New York, USA). Reagents and amino acids for making standard dropout (SD) plates for prototroph and colorimetric screening were obtained from Clontech.
pMD19T-Rheb and pMD19T-FKBP38 were generated in our previous work (Zheng et al., 2011;2012). The two plasmids were digested with restriction enzymes, respectively (TaKaRa, Dalian, China), to yield the Rheb and FKBP38 genes. The Rheb and FKBP38 genes were then cloned into pGADT7 to generate pGADT7-Rheb/FKBP38 (expressing the prey protein GAL4AD-Rheb/FKBP38), and pGBKT7 to generate pGBKT7-Rheb/FKBP38 (expressing the bait protein GAL4BD-Rheb/FKBP38), separately. The recombinant plasmids were verified by restriction analysis on a UV transilluminator (UVItec, Cambridge, UK) after electrophoresis and sequenced on an ABI PRISM 377XL DNA Sequencer (Applied Biosystems, Inc. Foster City, CA, USA).
In order to ensure the transformed yeast strains had a normal growth phenotype and were not been activated by single bait protein, the yeast strain yeast tow-hybird gold (Y2HGold) (Trp−) was transformed with the plasmids pGBKT7, pGBKT7-Rheb and pGBKT7-FKBP38, while the yeast strain Y187 (Leu−) was transformed with the plasmids pGADT7, pGADT7-Rheb and pGADT7-FKBP38, separately. For growth checking, the transformed yeast strain Y2HGoldpGBKT7, Y2HGoldpGBKT7-Rheb, and Y2HGoldpGBKT7-FKBP38 were inoculated on SD/-Trp plate and the Y187pGADT7, Y187pGADT7-Rheb and Y187 pGADT7-FKBP38 on SD/-Leu plate at a constant temperature of 30°C. For toxicity and self-active ability checking, the transformed yeast strain Y2HGold was cultured on SD/-Trp/X-α-Gal plate and examined with regard to bait protein expression, activation of transcription in the absence of a binding partner, and mating efficiency. These initial control studies are needed to ensure the goat Rheb or FKBP38 is effective bait in the screen.
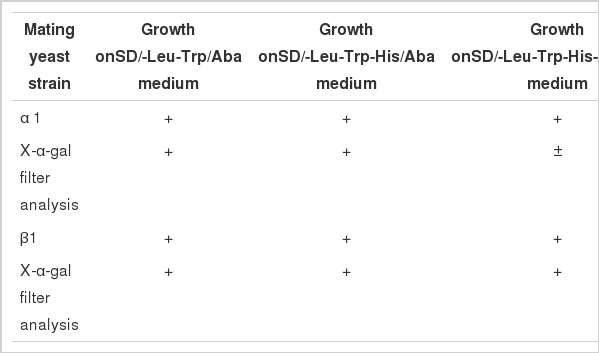
Goat Rheb or FKBP38 in the GAL4 activation domain of pGADT7, pretransformed into the yeast strain Y187, were screened by yeast two-hybrid assay by mating them to GAL4BD-FKBP38/Rheb-expressing Y2HGold on SD/-Leu-Trp (DDO) plate per the manufacturer’s instructions. Mating efficiency was determined to be within the acceptable limits per the manufacturer. Diploid cells were then screened on SD/-Leu/-Trp/Aba/X-α-Gal (DAX) plate, the blue colonies were transfered to SD/-Leu/-Trp/-His/X-α-Gal/Aba (HAX) plate and SD/-Leu/-Trp/-His/-Ade/Aba/X-α-Gal (QAX) plate for 3 to 6 days at 30°C, sequentially. Segregation was confirmed based on the appearance of white and blue clones in the process of subcultivation.
Statistical analysis
Descriptive statistics were generated for all quantitative data with presentation of mean±standard deviation.
RESULTS
Coimmunoprecipitation and western blot analysis
Rheb and FKBP38 were expressed in all normal and cotransfected cells. By western blot of transgenic GFb cells, cotransfected cells had an enhanced expression of Rheb and FKBP38, versus normal control GFb cells (Figure 1A).
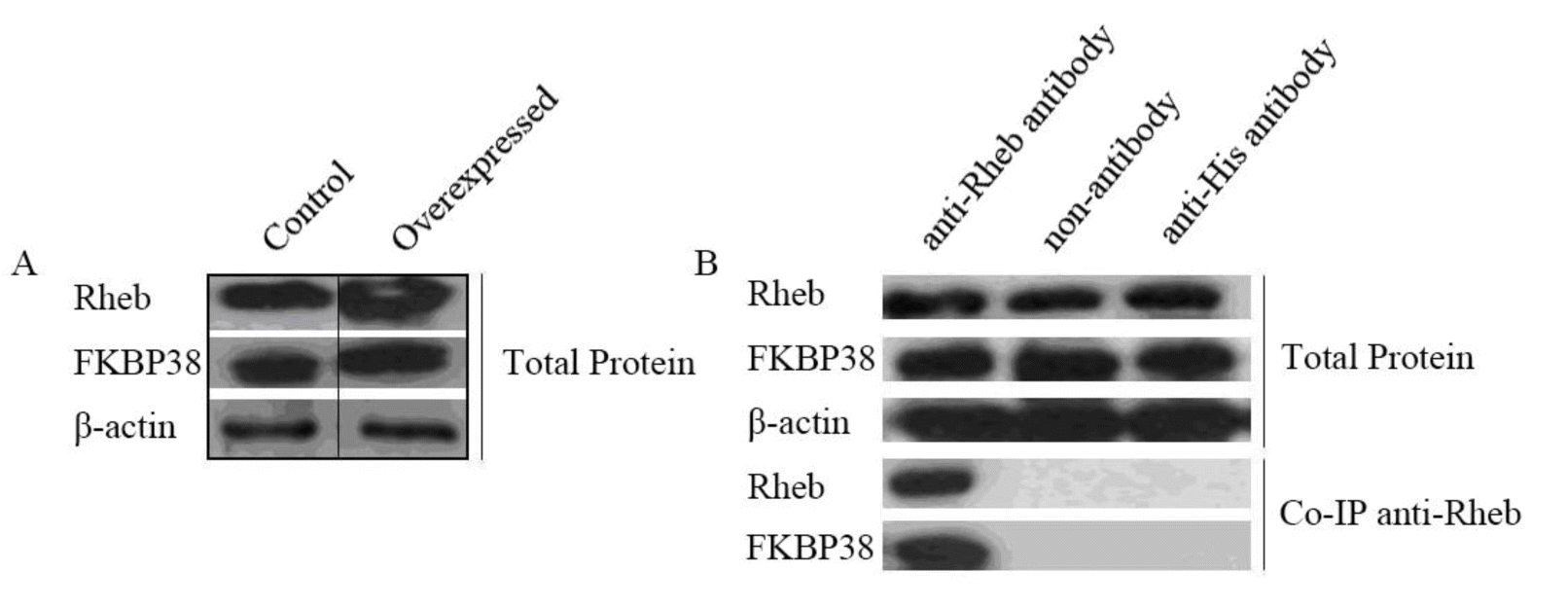
Western blot of total protein and co-IP with anti-Rheb. (A) Western blot analysis of total cell protein. Overexpressed cells had an enhanced expression of Rheb and FKBP38 versus normal control GFb cells; (B) co-IP analysis of eluates. Rheb and FKBP38 could be detected in anti-Rheb immunoprecipitates; β-actin is the internal control. Rheb, Ras homolog enriched in brain; co-IP, coimmunoprecipitation.
To examine the interaction between Rheb and FKBP38, we performed a co-IP experiment with anti-Rheb, observing that Rheb and FKBP38 were bound together in eluates from cotransfected cells (Figure 1B). The indicated two groups, no antibody and anti-His IgG, were used as in the control groups, which Rheb and FKBP38 were not detected while they could be detected in total proteins transgenic GFb cells and control groups; total proteins were used as a positive control. Our co-IP results demonstrate that Rheb and FKBP38 interact directly or indirectly.
Yeast mating
In order to exclude toxicity and self-activation of the bait proteins in Y2HGold cells, we transferred pGBKT7 and pGBKT7-Rheb as well as pGBKT7-FKBP38 into Y2HGold cells and pGADT7, pGADT7-Rheb and pGADT7-FKBP38 into Y187, separately. The cells grew normal on synthetic dropout nutrient medium SD/-Trp and SD/-Leu with a X-α-gal filter (Figure 2A), but the Y2HGold cell colonies containing pGBKT7-Rheb and pGBKT7-FKBP38 did not turn blue on the SD/-Trp/X-α-Gal (Figure 2B), suggesting that the indicated three plasmids and their encoded proteins were non-toxic and had no self-activation. The GAL4BD-Rheb and GAL4BD-FKBP38 are effective bait proteins in the screening.

Toxicity and self-activation of the bait proteins analyzed in Yeast cells (4×). (A) Phenotype for yeast transformation, 1, Y2HpGBKT7; 2, Y2HpGBKT7-Rheb; 3, Y2HpGBKT7-FKBP38; 4, Y187pGADT7; 5, Y187pGADT7-Rheb; 6, Y187 pGADT7-FKBP38. (B) Self-activation assay of Rheb and FKBP38. 1′ is Y2HGold with pGBKT7-Rheb and 2′ is Y2HGold with pGBKT7-FKBP38. Rheb, Ras homolog enriched in brain.
In order to verify the interaction between Rheb and FKBP38 complementary, the plasmids pGADT7-Rheb and pGADT7-FKBP38 for prey protein GAL4AD-Rheb and GAL4AD-FKBP38 were constructed. pGBKT7-Rheb and pGBKT7-FKBP38 were used to transform Y2HGold and the cells grew normally on SD/-Trp (Figure 2A-2,3) medium; meanwhile pGADT7-Rheb and pGADT7-FKBP38 were used to transform Y187 cells respectively and the cells grew normally on SD/-Leu medium (Figure 2A-5,6). Using Rheb and FKBP38 primers and the yeast colonies as templates, we amplified 555-bp (Rheb) and 1,236-bp (FKBP38) fragments (Figure 3). These results showed that the plasmids were transferred into yeast cells successfully.

PCR amplification of Rheb and FKBP38 using yeast colonies as templates; electrophoresis of 3 PCR products of (a) Rheb in yeast Y2H; (b) FKBP38 in yeast Y2H; (c) Rheb in yeast Y187; and (d) FKBP38 in yeast Y187; lane M: DL2000 marker. PCR, polymerase chain reaction; Rheb, Ras homolog enriched in brain.
In order to make the mating between the two yeast strains, the Y2HGold cells which contained pGBKT7-Rheb and Y187 cells which contained pGADT7-FKBP38 were mixed and cultured in yeast extract peptone dextrose medium (YPDA), while the Y2HGold cells which contained pGBKT7 and Y187 cells which contained pGADT7-FKBP38 were managed as control groups. The mating yeast strains were named as α1 and γ1, respectively (Figure 4). After 20 hours of mating and culture, clover-like diploid cells were observed by light microscopy, demonstrating that the yeast cells were able to grow on synthetic dropout nutrient medium (SD/-Leu-Trp [DDO]), suggesting successful yeast mating (Figure 4). At the same time, the reverse duplicate sets of experiments were managed, and the mating yeast strains were named as β1 and δ1, respectively.
Ras homolog enriched in brain and FK506 binding protein 38 coexist in yeast cells
In the next step of screening, γ1 and δ1 could not grow on synthetic dropout nutrient medium (SD/-Leu/-Trp/Aba/X-α-Gal [DAX]), suggesting that a single GAL4AD-Rheb/FKBP38 does not activate downstream reporter genes; thus, GAL4AD-Rheb and GAL4AD-FKBP38 could be used for further screening of yeast two-hybrid assay as prey protein. α1 and β1 grew and turned blue on SD medium (SD/-Leu/-Trp/Aba/X-α-Gal [DAX]) (Figure 5-1,2), indicating that the two reporter genes, AUR1-C and MEL1, were activated. The results implied that interactions probably existed between Rheb and FKBP38.
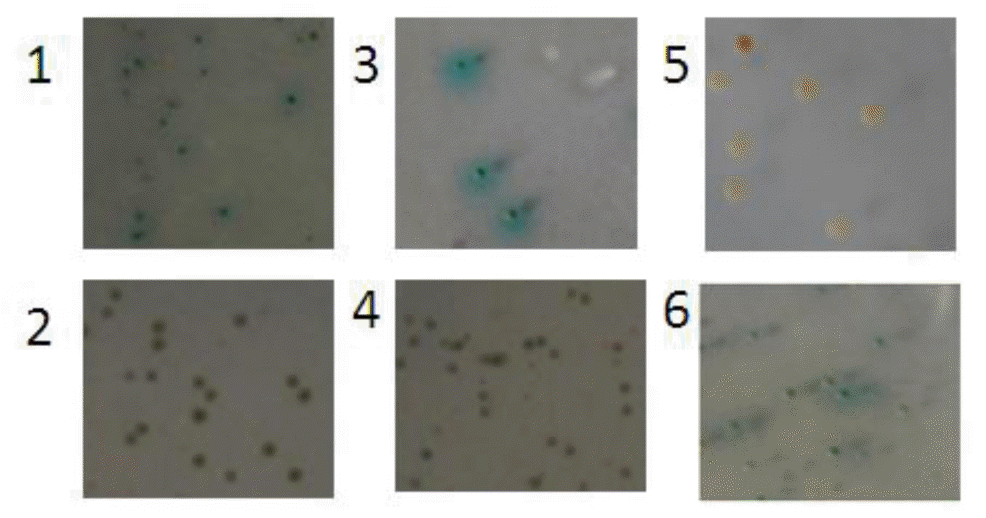
Colony screening using yeast two-hybrid system (4×). 1, α1 mating yielded blue colonies were selected on SD medium (SD/-Leu/-Trp/Aba/X-α-Gal [DAX]). 2, β1 mating yielded blue colonies on SD medium (SD/-Leu/-Trp/Aba/X-α-Gal [DAX]). 3, Blue yeast colonies of α1 mating yeast strain were selected on SD medium (SD/-Leu/-Trp/-His/X-α-Gal/Aba [HAX]). 4, Blue yeast colonies of β1 mating yeast strain were selected on SD medium (SD/-Leu/-Trp/-His/X-α-Gal/Aba [HAX]). 5, α1 grew white colonies on SD medium (SD/-Leu/-Trp/-His/-Ade/Aba/X-α-Gal [QAX]). 6, Blue yeast colonies of β1 mating yeast strain were selected in the fourth round on SD medium (SD/-Leu/-Trp/-His/-Ade/Aba/X-α-Gal [QAX]). SD, standard dropout.
The blue colonies of α1 and β1 were then transferred and cultured on SD/-Leu/-Trp/-His/X-α-Gal/Aba (HAX) plate. α1 and β1 yeast cells were able to grow on the plate (Figure 5-3,4). On the fourth day, the robust blue colonies were replanted on a 9-cm SD/-Leu/-Trp/-His/-Ade/Aba/X-α-Gal (QAX) agar plate for further screening. The results showed that β1 cells were able to grow on SD/-Leu/-Trp/-His/-Ade/Aba/X-α-Gal (QAX) medium with a positive X-α-gal filter, which indicated that Rheb and FKBP38 interacted (Figure 5-6) with Rheb as prey protein and FKBP38 as bait protein. Meanwhile, α1 cells were able to grow on SD medium (SD/-Leu/-Trp/-His/-Ade/Aba/X-α-Gal [QAX]) with a negative X-α-gal filter, indicating that Rheb and FKBP38 interact weakly with Rheb as bait protein and FKBP38 as prey protein (Figure 5-5). Based on the experiments above, we detected an interaction between Rheb and FKBP38 and listed the possible interactions in Table 1.
DISCUSSION
Rheb belongs to the Ras family, which encodes a carboxyl-terminal CAAX box, suggesting that the protein undergoes posttranslational farnesylation—the membrane localization of Rheb through farnesylation is important for upstream regulation of mTOR signaling (Basso et al., 2005; Hanker et al., 2010). As a unique member of the FKBP family, FKBP38 contains three types of domains: FKBP-type peptidyl-prolyl cis-trans isomerase (FKBP_C), tetratricopeptide repeat domain (TPR). In the absence of growth factors and nutrition, the FKBP_C domain can bind to mTOR to restrain the phosphorylation of S6K1 and 4EBP1, which are downstream regulatory factors of mTOR signaling (Bai et al., 2007).
The products of cashmere goat Rheb gene and FKBP38 gene were characterized by informatics in our previous study. Goat Rheb has a RAS domain (consisting of switch I and switch II domains) from residues 4 to 170 and a carboxyl-terminal CAAX box (Zheng et al., 2011). Goat FKBP38 contains an FKBP_C domain, 2 TPR domains, and a TM domain and CAAX box (Zheng et al., 2012). These features are similar to those of Rheb and FKBP38 in other species, indicating that the deduced goat Rheb protein has Ras-like small GTPase activity and that FKBP38 has intrinsic FKBP family activity.
Rheb is a Ras-like small GTPase that shuttles between an active GTP-bound form and an inactive GDP-bound form. Rheb interacts directly with mTOR and activates mTOR kinase (Yadav et al., 2013), whereas Rabin8 interacts with Rheb and inhibits the phosphorylation of Ser235/Ser236 in small ribosomal subunit protein S6 (Parkhitko et al., 2011). Rheb is also regulated through direct phosphorylation by MAP kinase-activated protein kinase 5, and Rheb-mediated mTORC1 activation is inhibited (Zheng et al., 2011); cyclic adenosine monophosphate can control mTOR activation by regulating the dynamic interaction between Rheb and phosphodiesterase 4D (Kim et al., 2011), and the N-methyl-D-aspartate receptor subunit N-methyl-D-aspartate 3A can interact with Rheb to function as a break of mTOR-dependent synaptic translation (Sucher et al., 2010).
FKBP38 belongs to the FKBP family and was believed to have PPIase activity. However, unlike other characterized members of the FKBP family, FKBP38 might not possess such activity, because its FKBP_C domain contains several mutations in the PPIase active site (Lam et al., 1995; Liu et al., 2003; Weiwad et al., 2005). In addition, mutants without the TM domain are unable to associate with mitochondria, causing FKBP38 to lose its ability to inhibit mTOR, suggesting that mitochondrial localization is essential for FKBP38 to inhibit mTOR signaling (Ma et al., 2008). Conversely, it appears that the CAAX box is linked to mitochondrial localization (Shirane et al., 2003)—the mitochondrial localization of FKBP38 is important for its activation and interaction with other proteins.
In the current model of the Rheb-FKBP38-mTOR relationship, FKBP38 is an endogenous inhibitor of mTOR. Under amino acid or serum starvation, this mTOR inhibitor binds to and interferes with mTORC1 function similarly to the FKBP12-rapamycin complex. Under conditions of ample growth factors and nutrients, the FKBP_C domain might interact with Rheb-GTP to activate downstream mTOR signaling by releasing mTOR from FKBP38 and the Switch I domain of the Rheb-mediated interaction with FKBP_C. Rheb, FKBP38, and mTOR coexist on mitochondria, and their interaction might depend on their localization in living cells (Ma et al., 2008). This mechanism likely explains the results in which the interaction between Rheb and FKBP38 is undetectable in vitro (Uhlenbrock et al., 2009).
In this study, we overexpressed goat Rheb and FKBP38 in goat fetal fibroblasts, extracted whole proteins, and performed co-IP to detect them by western blot. We found Rheb binds directly to FKBP38. Then, we constructed bait vectors (pGBKT7-Rheb/FKBP38) and prey vectors (pGADT7-Rheb/FKBP38), and examined their interaction by yeast two-hybrid assay. Their direct interaction was observed, regardless of which plasmid served as the prey or bait vector. These results indicate that the 2 proteins interact directly in vivo.
CONCLUSION
Collectively, we have demonstrated the direct interaction between Rheb and FKBP38 in Cashmere goat cells, providing new evidence on the relationship between Rheb and FKBP38. Our results add to the genetic and functional data on Rheb and FKBP38 and clarify the function and mechanism of the mTOR signaling pathway.
ACKNOWLEDGMENTS
This work was supported by a grant from the National natural Sciences Foundation of China (No. 31160469, 31360561) and Natural Sciences Foundation of Inner Mongolia (2011MS0521), a graduate student research project of Inner Mongolia University, and Major Projects for New Varieties of Genetically Modified Organisms (No.2014ZX08008-002).