Genome-wide Single Nucleotide Polymorphism Analyses Reveal Genetic Diversity and Structure of Wild and Domestic Cattle in Bangladesh
Article information
Abstract
In spite of variation in coat color, size, and production traits among indigenous Bangladeshi cattle populations, genetic differences among most of the populations have not been investigated or exploited. In this study, we used a high-density bovine single nucleotide polymorphism (SNP) 80K Bead Chip derived from Bos indicus breeds to assess genetic diversity and population structure of 2 Bangladeshi zebu cattle populations (red Chittagong, n = 28 and non-descript deshi, n = 28) and a semi-domesticated population (gayal, n = 17). Overall, 95% and 58% of the total SNPs (69,804) showed polymorphisms in the zebu and gayal populations, respectively. Similarly, the average minor allele frequency value was as high 0.29 in zebu and as low as 0.09 in gayal. The mean expected heterozygosity varied from 0.42±0.14 in zebu to 0.148±0.14 in gayal with significant heterozygosity deficiency of 0.06 (FIS) in the latter. Coancestry estimations revealed that the two zebu populations are weakly differentiated, with over 99% of the total genetic variation retained within populations and less than 1% accounted for between populations. Conversely, strong genetic differentiation (FST = 0.33) was observed between zebu and gayal populations. Results of population structure and principal component analyses suggest that gayal is distinct from Bos indicus and that the two zebu populations were weakly structured. This study provides basic information about the genetic diversity and structure of Bangladeshi cattle and the semi-domesticated gayal population that can be used for future appraisal of breed utilization and management strategies.
INTRODUCTION
Livestock are an integral part of the Bangladeshi agricultural economy. About 12% of the agricultural GDP comes from the livestock sector, and 10 million people are directly involved with the livestock sector for their livelihoods (Karim et al., 2010). In Bangladesh, cattle are the most versatile among livestock species in the existing integrated agricultural farming system. There are approximately 24.5 million heads of cattle in Bangladesh, which comprise approximately 1.79% of the world cattle population and 5.47% of the Asian cattle population (Baker, 2004). Indigenous cattle breeds and types are better adapted to adverse climatic conditions, poor nutrition, and low management systems, and they are more resistant to local diseases and parasites. Bangladeshi cattle are zebu type, and they are identified by their local name or the place where they predominantly found, such as Pabna, red Chittagong (RC), Munshiganj, Madaripur, and north Bengal grey. The commonly available native types are called non-descript deshi (ND). Indigenous Bangladeshi cattle populations have lower productivity than the improved crossbred (e.g. Shahiwal, Sindhi and Holstein-Friesian) animals. There is also variation in coat color, size, live weight, and production and reproduction traits. However, genetic differences among these populations are not well understood. Gayal (Bos frontalis) is a large, semi-domesticated bovine species seen in the southeastern hilly regions of Bangladesh. Studies have shown species hybridization between gayal and the local cattle (B. indicus) (Giasuddin et al., 2003).
Among the wide range of molecular markers developed, single nucleotides polymorphisms (SNPs) are the most abundant, are widely dispersed throughout genomes, and have variable distribution among species (Vignal et al., 2002). The availability of high-throughput SNP genotyping platforms makes it feasible to undertake high-density scans by using large numbers of SNP markers and are either distributed across the whole genome or focused in specific regions. The SNPs are useful in studying livestock genetic diversity and population structure (McKay et al., 2008; Lin et al., 2010). Although a large number of SNPs have been identified from the bovine genome-sequencing project, few of these have been validated, particularly in B. indicus breeds. Therefore, we used high density bovine SNP 80K to assess genetic diversity and population structure of Bangladeshi indigenous cattle populations and the semi-domesticated gayal.
MATERIALS AND METHODS
Populations and DNA sample collection
Among phenotypically categorized indigenous Bangladeshi zebu cattle breeds, we collected nasal samples from unrelated RC (n = 28) and ND (n = 28) animals. The two populations are notably different in their distinct coat color types (Bhuiyan et al., 2007a). The RC samples were collected from the Chittagong district, which is the only breeding area for this population. The ND samples were collected from Mymensingh region, but they had wide distribution (Bhuiyan et al., 2007b). For comparison, we also collected 17 samples from the gayal population of the Bandarban district, a hilly region of Bangladesh. Nasal samples were collected using Performagene Livestock’s nasal swab DNA collection kit and DNA was extracted from nasal samples according to the manufacturer’s recommendations (DNA Genotek Inc., 2012).
Genotyping and marker selection
All samples were genotyped using the GeneSeek Genomic Profiler Indicine HD Beadchip, an Illumina Infinium array consisting of nearly 80,000 SNPs derived mainly from B. indicus breeds (GeneSeek, Lincoln, NE, USA), according to Illumina’s standard protocols (http://www.illumina.com). For minor allele frequency (MAF) estimation, we analyzed approximately 69,804 autosomal SNP markers. For diversity analysis, SNPs were screened based on the following criteria: call rate ≥95, MAF ≥5%, and Hardy-Weinberg equilibrium, HWE ≥0.001. The diversity analysis resulted in 51,366 SNPs. To save running time in the population structure analysis, 35,437 SNPs were screened based on the following criteria: call rate ≥95, MAF ≥20%, and HWE ≥0.001.
Statistical analysis
Minor allele frequency and proportion of polymorphic SNPs were estimated using the Golden Helix SNP Variation Suite software version 7 (Golden Helix, 2013). Within breed genetic variability (observed and expected heterozygosity) and inbreeding (Weir, 1996) estimates were calculated using PowerMarker V3.25 software (Liu and Muse, 2005). Reynolds genetic distances (Reynolds et al., 1983) between pairs of populations were estimated using the same software. Population genetic structure was inferred by applying principal component analysis (PCA) using the Golden Helix SNP Variation Suite software version 7 (Golden Helix, 2013) for the whole data set and by model-based clustering using STRUCTURE 2.3.4 (Prichard et al., 2000). The PCA estimates were performed for the three studied populations by using the allele frequencies of 69,804 SNP markers. We ran STRUCTURE following the admixture ancestry model and correlated allele frequency for K values of 2 and 3 with a burn period of 20,000 generations and Markov chain Monte Carlo simulations of 100,000 iterations using the correlated allele model.
RESULTS
Minor allele frequency and genetic diversity
The mean MAFs and within population genetic variations are presented in Table 1. Common SNP variants with MAFs ≥0.10 and ≤0.50 ranged from 30.31% in gayal to 88.57% in ND populations. More than half of the SNPs (52.48%) in RC and ND displayed MAFs greater or equal to 0.30, whereas only 7.72% of SNPs in gayal were in this range. On average, 95.18% of SNPs (69,804) displayed polymorphisms (MAF≥0.05) within the B. indicus populations (RC and ND). However, only 57.73% of SNPs showed polymorphisms in gayal. The observed heterozygosity was 0.415±0.143 and 0.420±0.143 in RC and ND, respectively (Table 2). Gayal had the lowest observed and expected heterozygosities (0.148±0.143 and 0.153±0.139, respectively) and a significant heterozygosity deficiency (0.061±0.229 [FIS]). The estimated FIS values in RC and ND were −0.029±0.192 and −0.027±0.206, respectively (Table 2). Gayals had a larger proportion of monomorphic SNPs, which accounted for 79.31% of monomorphic SNPs shared with B. indicus (RC and ND) (Table 3). However, approximately 60% of polymorphic SNPs were shared by gayal and the B. indicus population. High differences in the number of monomorphic SNPs were found on chromosome 1, wherein 152 SNPs were common and accounted for 85.88% of the monomorphic SNPs on the same chromosome. Less difference were found on chromosome 27, wherein only 38 monomorphic SNPs were common and accounted for 67.86% of the monomorphic SNPs. Almost all of the polymorphic SNPs on gayal chromosomes were also found to be polymorphic in the zebu population, which may reflect a high deficiency in heterozygosity observed in the gayal population.

Numbers of polymorphic (MAF ≥0.05) and monomorphic (MAF <0.05) SNP markers in domestic cattle (ND and RC) and gayal populations across chromosomes
Genetic differentiations between the two Bangladeshi zebu populations were very low, and approximately 99% of the genetic variation was retained within the breed. Similarly, genetic distance (Reynolds distance) showed a close relationship between RC and ND (0.020). With an FST value of 0.33 and Reynolds distances of 0.31, gayal showed strong differentiation from B. indicus subspecies.
Population structure
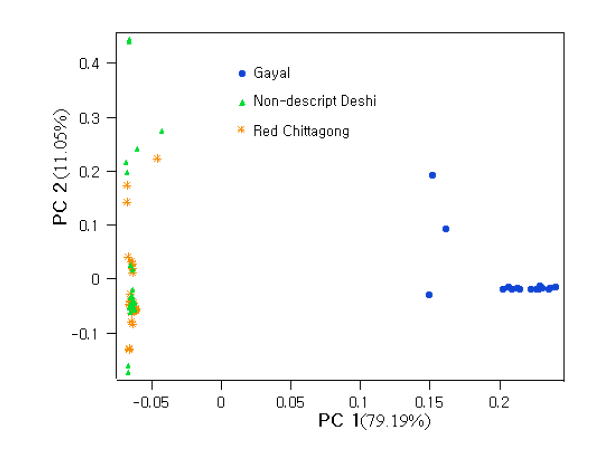
The first and the second principal components (PC1 and PC2) explained 90.24% of the total variation and evidently distinguish the two zebu populations from gayal. The results coincided well with the STRUCTURE output at K = 2 and K = 3 (Figure 2). The output at K = 2 seems plausible; it clearly distinguished the gayal from B. indicus populations (RC and ND) with some level of gene flow between the breeds. The output at K = 3 suggests higher admixture than expected in ND and RC, which does not agree with the PCA result in this study. This might be because of the influence of exotic blood due to indiscriminate crossbreeding. Approximately 11% of the gayal population is considered to share common ancestry with the ND and RC populations.
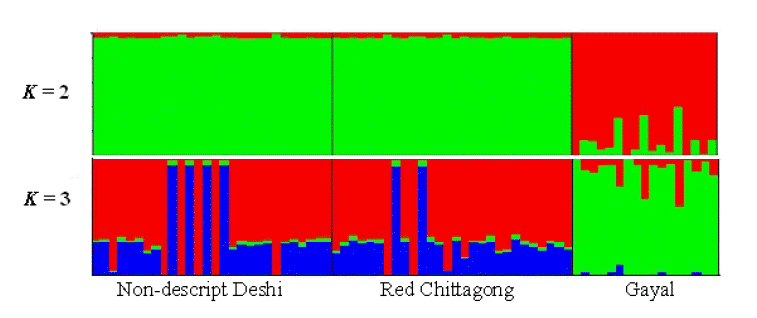
Genetic structure of three cattle populations inferred using structure 2.3.4 (Pritchard et al., 2000). Each individual animal is represented using a single vertical line divided by k colors, where k is the number of clusters assumed and the length of the colored segment represents the individual’s estimated proportion of membership to a particular cluster.
DISCUSSION
Evaluation of genetic diversity and assessment of population structure is necessary to appraise the utilization and management of farm animal genetic resources. In this study, we estimated genetic diversity and population structure of Bangladeshi zebu cattle populations and the semi-domesticated gayal breed by using a high-density SNP genotyping chip recently developed from indicine cattle. The indigenous zebu cattle of Bangladesh have been reared using traditional husbandry practices, and intensive selective breeding, for milk or meat, is rare. The practices result in an effective population size that is much larger than the recognized breed. The attributes contributed to the high diversity measures in Bangladeshi zebu cattle. The low genetic differentiation observed between RC and ND could be because of their common origin and gene flow. In addition, it is possible that a spontaneous mutation or aggregation of genes for coat-color differentiated RC occurred from ND and has been maintained by the farmers of the greater Chittagong district. However, apart from this phenotypic feature, there is no remarkable difference between the two zebu populations in production traits.
The zebu cattle MAFs that we found were higher than that reported for Ethiopian cattle populations (Edea et al., 2013) and for B. indicus and B. taurus breeds (Lin et al., 2010). Variation across breeds may be because of the difference in the chip employed. The higher MAF reported here might be because the SNP panels that we used were derived from B. indicus breeds, and as expected, the minor allele was higher in the zebu breeds. The level of polymorphism we found was higher than that shown in previous studies of taurine (95.21%) and African cattle populations (83.96%) (Edea et al., 2013). As Bangladeshi zebu cattle are non-selected random bred indigenous population, expectedly higher heterozygosity remains over the generations, and therefore, the observed and expected heterozygosities were higher than those reported for African cattle populations (Edea et al., 2013) and taurine breeds (Lin et al., 2010).
A total of 29,496 SNP markers were monomorphic (MAF<0.05) in the gayal population, whereas the total number of monomorphic SNP makers was 3364 in the zebu population (Table 3). Interestingly, approximately 79.31% of monomorphic markers in the zebu population were also monomorphic in the gayal population, which might indicate an influence of zebu on gayal. This result supports the hypothesis that gayal are hybrid descendants of wild gaurs and domestic cattle of either B. indicus or B. taurus (Payne, 1970). Species hybridization between gayal and local cattle (B. indicus) has been reported (Giasuddin et al., 2003). In Bangladesh, some tribal family rear gayal with native cattle and their hybrids are sometimes found in local markets. The total number of polymorphic (MAF≥0.05) SNPs screened were 40,299 and 66,440 in gayal and zebu populations, respectively. Regardless of the effect of origin of the chip, the high genome-wide number of monomorphic SNPs in the gayal population could be attributed to the FIS of 0.06. The Gayal population has been declining over time, which has led to the reduced effective population size in the region. The observed variation of SNPs between the two subspecies should be studied further to increase the understanding of the genetics of the phenotypic differences. The candidate gene approach could be used to determine the role of specific genes in these bovine species.
The PCA results clustered the RC and ND as one population, whereas the gayal population was observed as a separate cluster (Figure 1). Within the gayal cluster, a few were outgroups. This is likely because a few hybrid individuals were noted within the studied gayal population. The structure output also revealed the same genetic architecture in the two zebu populations, with some introgressions of genes from domestic cattle (i.e., zebu) to the gayal population and vice versa. This might be due to inter-specific hybridization among the Bos species (zebu, taurine, gayal, and yak) which has been documented in South and Southeast Asian cultures (Tanaka et al., 2011). The admixed zebu and gayal individuals in the studied population might be due to sampling errors despite they have the phenotypic resemblance and every precautions were taken during sampling. It is notable that the remnant of inheritance in admixed individuals is very difficult to trace phenotypically in some cases. In addition, the pedigree information is almost unknown to farmers in Bangladesh perspective. The bottleneck of genetic features in the gayal population might be because of inbreeding within a small population. In fact the geographic distribution of gayal habitats is very specific where people of several tribal families used to keep their animals together in a form of small group in the particular territory. Random mating occurs in that small sub-population over the years despite hierarchy of breeding bulls is a question. Moreover, the chance of interbreeding among the sub-population is negligible. This faulty breeding practices and lack of scientific management leads to increase homogeneity within the small group. However, our attempts unveiled the genetic architecture of gayal for the first time and above all, gave us hints to take immediate attention from different scientific institutes of Bangladesh to overcome the bottlenecks.
ACKNOWLEDGMENTS
This study was designed by a Memorandum of Understanding agreement between Chungbuk National University and Bangladesh Agricultural University, and was supported by a grant from Next Generation Bio-green 21 Program under Rural Development Administration.