 |
 |
Abstract
The A-type lamin deficient mouse line (LmnaŌłÆ/ŌłÆ) has become one of the most frequently used models for providing insights into many different aspects of A-type lamin function. To elucidate the function of Lmna in the growth and metabolism of mice, tissue growth and blood biochemistry were monitored in Lmna-deficient mice, heterozygous (Lmna+/ŌłÆ) and wide-type (Lmna+/+) backcrossed to C57BL/6 background. At 4 weeks after birth, the weight of various organs of the LmnaŌłÆ/ŌłÆ, Lmna+/ŌłÆ and Lmna+/+ mice was measured. A panel of biochemical analyses consisting of 15 serological tests was examined. The results showed that Lmna deficient mice had significantly decreased body weight and increased the ratio of organ to body weight in most of tissues. Compared with Lmna+/+ and Lmna+/ŌłÆ mice, LmnaŌłÆ/ŌłÆ mice exhibited lower levels of ALP (alkaline phosphatase), Chol (cholesterol), CR (creatinine), GLU (glucose), HDL (high-density lipoprotein cholesterol) and higher levels of ALT (alanine aminotransferase) (p<0.05). LmnaŌłÆ/ŌłÆ mice displayed higher AST (aspartate aminotransferase) values and lower LDL (lowdensity lipoprotein cholesterol), CK-MB (creatine kinase-MB) levels than Lmna+/+ mice (p<0.05). There were no significant differences among the three groups of mice with respect to BUN (blood urea nitrogen), CK (creatine kinase), Cyc C (cystatin C), TP (total protein), TG (triacylglycerols) and UA (uric acid) levels (p>0.05). These changes of serological parameters may provide an experimental basis for the elucidation of Lmna gene functions.
Lamins are intermediate filament proteins in the nuclear lamina and the cellular matrix and are critical determinants of nuclear architecture. They are important regulators of gene expression through their involvement in signaling, transcription, and chromatin organization (Andres and Gonzalez, 2009; Mendez-Lopez and Worman, 2012). Mutations in Lmna are responsible for more than ten different disorders, commonly referred to as ŌĆ£laminopathiesŌĆØ. These diseases are characterized by tissue-specific (striated muscle, adipose tissue, peripheral nerve) or systemic effects (premature ageing syndromes). Lmna mutations cause autosomal-dominant Emery-Dreifuss muscular dystrophy (EDMD) and related diseases with dilated cardiomyopathies (DCM) that affect cardiac muscle and skeletal muscle to variable degrees (Bonne et al., 1999; Raffaele Di Barletta et al., 2000). Other Lmna mutations affect adipose tissue or peripheral neurons, causing Dunnigan-type familial partial lipodystrophy (FPLD) (Cao and Hegele, 2000; Worman et al., 2010; Bong Hwan Choi, 2012) or Charcot-Marie-Tooth type 2B1 disease (De Sandre-Giovannoli et al., 2002), respectively. Some Lmna mutations cause progeroid syndromes, which are characterized by features of accelerated aging (Eriksson et al., 2003; Arancio, 2012). Remarkable progress has been made in unraveling the molecular pathophysiology underlying these disorders. However, the mechanism whereby different mutations in Lmna cause such diverse diseases remains a puzzle.
The first genetic model for the study of laminopathy was the Lmna knockout mouse, which has become one of the most frequently used models to determine relationships between physiological functions of nuclear lamins and their contribution to the molecular pathophysiology of laminopathies. This mouse model has provided profound insights into many different aspects of A-type lamin functions. These Lmna knockout mice appear normal at birth but develop severe muscular dystrophy and die within eight weeks (Jahn et al., 1999; Sullivan et al., 1999). In the first reported human Lmna-null case, a baby possessing a homozygous nonsense mutation in Lmna, with no detectable A-type lamin proteins, died after premature birth, exhibiting similar clinical features, severe joint contractures, muscular dystrophy, fibrosis and the absence of muscle fibers in the diaphragm (Muchir et al., 2003). The resemblance of the babyŌĆÖs symptoms to conditions observed in laminopathic mice suggests that A-type lamins, although not essential for cell viability, are essential for human life.
Different mutations in Lmna can produce different diseases, involving the skeleton, muscle, fat and many types of cell and tissue abnormalities, prompting lamin A/C functional diversity. The Lmna gene deletion in mice is one of the most versatile animal models for lamin function, providing insights into various issues of A-type lamin function. How the Lmna gene functions and why gene deletions cause the observed bodily changes are not entirely clear. In the diagnostic process of many clinical diseases, a patientŌĆÖs serum constituents are measured to evaluate specific organ functions, including liver function, heart function, and/or renal function. These results will aid in the physicianŌĆÖs diagnosis of disease. This article explores the changes in growth and serum biochemistry in C57BL/6 mice due to homozygous or heterozygous deletion of the Lmna gene. These changes of serological parameters can implicate the affected organs of mice, and provide an experimental basis for the elucidation of Lmna gene functions.
Lmna knockout mice in a C57BL/6 genetic background were generated as described previously (Sullivan et al., 1999). Lmna+/ŌłÆ mice were kindly provided by Professor Collin Stewart and were used to establish a heterozygous breeding colony of mice with a null allele of Lmna to obtain LmnaŌłÆ/ŌłÆ, Lmna+/ŌłÆ and Lmna+/+ (n = 10 per group). The mice were genotyped by PCR using genomic DNA obtained from the tail using the Manual Extraction DNA Purification Kit (Beyotime Institute of Biotechnology, China) with Lmna wild-type and mutant allele primers. The sequences of Lmna wild-type primers were 5ŌĆ▓-GCT GAG TAC AAC CTG CGC TCA C-3ŌĆ▓ and 5ŌĆ▓-GTA GGA GCG GGT GAC TAG GTT G -3ŌĆ▓. The sequences of the Lmna mutant allele primers were 5ŌĆ▓-TGT GGA ATG TGT GCG AGG CCA G -3ŌĆ▓ and 5ŌĆ▓-GTA GGA GCG GGT GAC TAG GTT G -3ŌĆ▓. The target fragments were amplified for 40 cycles with denaturing at 95┬░C for 40 s, annealing at 62┬░C for 40 s and extending at 72┬░C for 60 s.
Experimental procedures and mice breeding procedures were performed as stipulated by the guidelines on animal research of the Animal Research Ethic Committee of the Guangdong Medical College, China. Animal care personnel paired one male and one female mouse in individually-ventilated microisolator cages. Each morning, females were checked for a vaginal plug as a sign of mating. Plug-positive females were individually housed throughout gestation and were allowed to develop their pregnancies to term. At 3 weeks of age, offspring were weaned from their mothers, and the sexes caged separately. Up to five sibling mice were housed per cage after weaning. Each cage housing a breeding unit pair was allowed free access to Purina Rodent Chow and tap water ad libitum. They were maintained in a controlled environment at 25┬▒3┬░C and 75┬▒2% relatively humidity with a 12:00/12:00-h dark/light cycle.
The mice were divided into three groups according to their determined genotype. Each group contained 10 male mice, and the LmnaŌłÆ/ŌłÆ group contained an additional 10 female mice. Blood samples were obtained by retro-orbital phlebotomy, and the mice were sacrificed at 4 weeks of age. Blood was collected into serum tubes between 09:00 and 11:00 a.m. After 30 min incubation at room temperature, the samples were centrifuged for 10 min at 2,500 rpm to obtain the serum at 4┬░C. We observed only a few cases of hemolysis, which were excluded from the study. The isolated serum was stored at ŌłÆ80┬░C until analysis.
Fifteen parameters were measured from a single serum sample using an Olympus AU640 autoanalyzer (Olympus, Hamburg, Germany) and adapted reagents from Olympus (Hamburg, Germany) at 37┬░C. Calibration and quality control of the machine were performed daily according to the manufacturerŌĆÖs protocols and using the calibration samples obtained from the manufacturer. The clinical chemistry screen included the following parameters: i) the glycolipid panel including GLU, TG, Chol, HDL and LDL; ii) the heart panel including CK and CK-MB; iii) the liver panel including ALT, AST, ALP, and TP; iv) the kidney panel including BUN, Cyc C, Cr, and UA.
The body weights of LmnaŌłÆ/ŌłÆ, Lmna+/+ and Lmna+/ŌłÆ mice were observed once every 3 days beginning at birth (D1) and ending 4 weeks after birth (D28). The weights of various organs, including the brain, heart, liver, kidney, lung, spleen, thymus gland, testis and left gastrocnemius muscle, were also determined at the end of the growth observation, and the organs were stored in liquid nitrogen. Tissues were removed free of adhering tissue and washed with normal saline after the blood was collected from the mice. Excess moisture was absorbed by clean filters, and the tissues were weighed immediately.
Statistical analyses were performed with SPSS (v13.0). Mean values and standard deviations were calculated for each parameter. The results are expressed as the mean┬▒SD, and differences between groups of mice were determined by LeveneŌĆÖs t-test, the Satterthwaite t-test, the Student-Neuman-Keuls (SNK) test, or DunnettŌĆÖs T3 test. P values less than 0.05 were considered statistically significant.
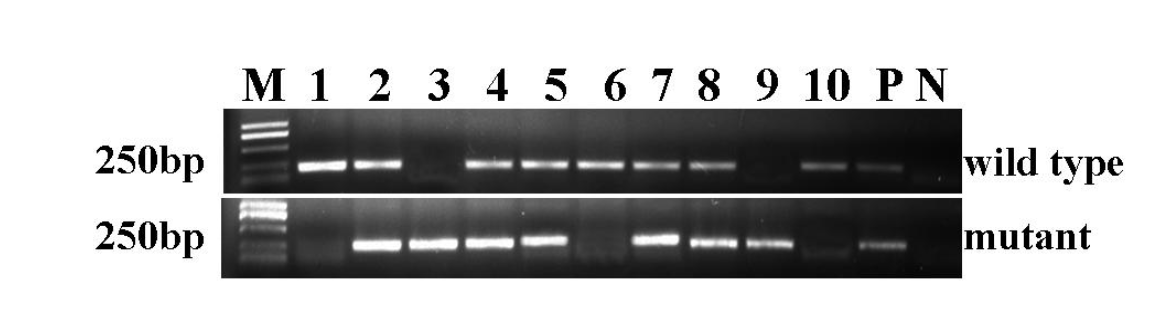
The polymerase chain reaction (PCR) was used to verify the genotype of the mice using two pairs of primers specific to the sequences in wild-type and mutant Lmna. The wild-type mice exhibited a band of 210 bp, heterozygous mice exhibited bands at both 210 bp and 230 bp, and a single band of 230 bp was observed in homozygous Lmna deletion mice. In total, 117 mice from 13 litters were tested, among which 29 were confirmed to be wild-type, 21 were homozygous and the remainder were heterozygous. This was roughly consistent with the proportions expected based on the principle of Mendelian inheritance regularity. Figure 1 shows a representative genotyping of a 10-mouse litter by electrophoresis. Mice No. 1, No. 6, and No. 10 were wild-type. No. 3 and No. 9 were homozygous for the Lmna deletion. The rest were heterozygous for the deletion of Lmna.
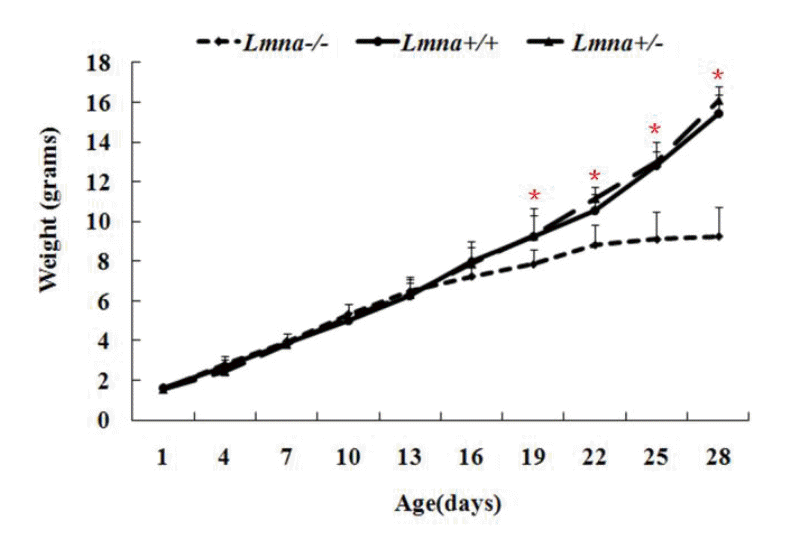
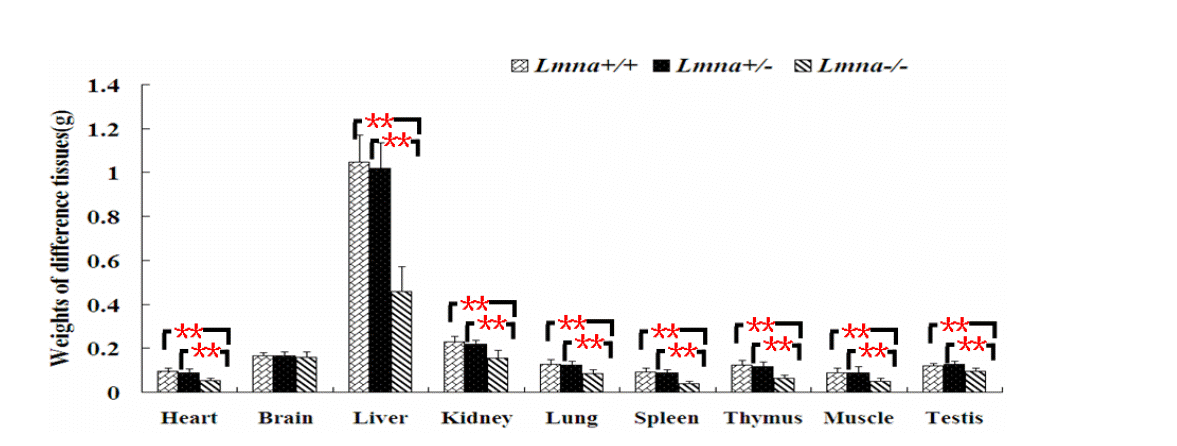
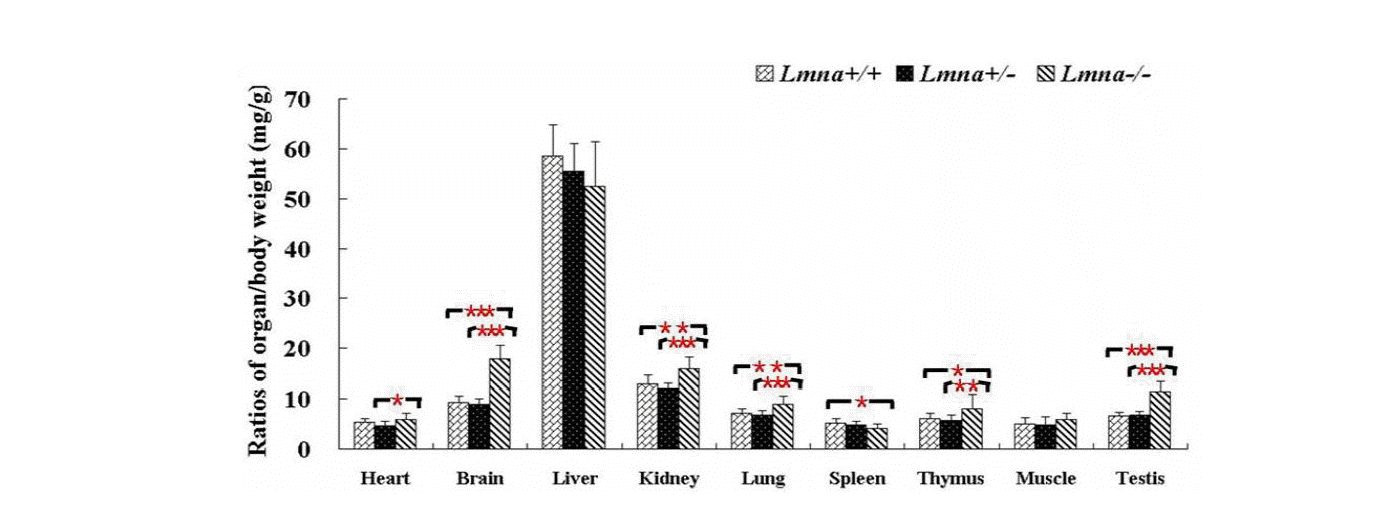
To examine the effects of Lmna deficiency on mouse growth, we measured the body weights from Lmna+/+, Lmna+/ŌłÆ and LmnaŌłÆ/ŌłÆmice. The results showed that LmnaŌłÆ/ŌłÆ mice had significantly decreased body weight compared with Lmna+/+ and Lmna+/ŌłÆ mice (Figure 2). At 28 days after birth, the body weight of LmnaŌłÆ/ŌłÆ mice were about one half of Lmna+/+ and Lmna+/ŌłÆ mice, while no significant differences in the growth of Lmna+/+ and Lmna+/ŌłÆ mice were observed. The tissue weights and the organ to body weight ratios from mice with different genotypes are shown in Figure 3 and Figure 4. Compared with Lmna+/+ and Lmna+/ŌłÆ mice, the brain to body weight ratios increased dramatically (p<0.05) in LmnaŌłÆ/ŌłÆ mice, although there are no significant differences (p>0.05) in the weights of the brain tissue from the three groups. In contrast, the organ (lung, kidneys, thymus and testis) to body weight ratio increased drastically (p<0.05), although the absolute weight of four organs decreased (p<0.05) in LmnaŌłÆ/ŌłÆ mice at 4 weeks of age. LmnaŌłÆ/ŌłÆ mice also had an elevated ratio for their muscle and hearts, while there were only significant differences between the heart to body weight ratio found in Lmna+/ŌłÆ and LmnaŌłÆ/ŌłÆ mice (p<0.05). A similar decrease in the weight of the lung and spleen (p<0.05), but with a decrease in organ: body weight ratio as well, there were only significant differences between the spleen: body weight ratio found in Lmna+/+ and LmnaŌłÆ/ŌłÆ mice (p<0.05). There were no significant differences between the tissue weights and the organ: body weight ratio found in Lmna+/+ and Lmna+/ŌłÆ mice (p>0.05).
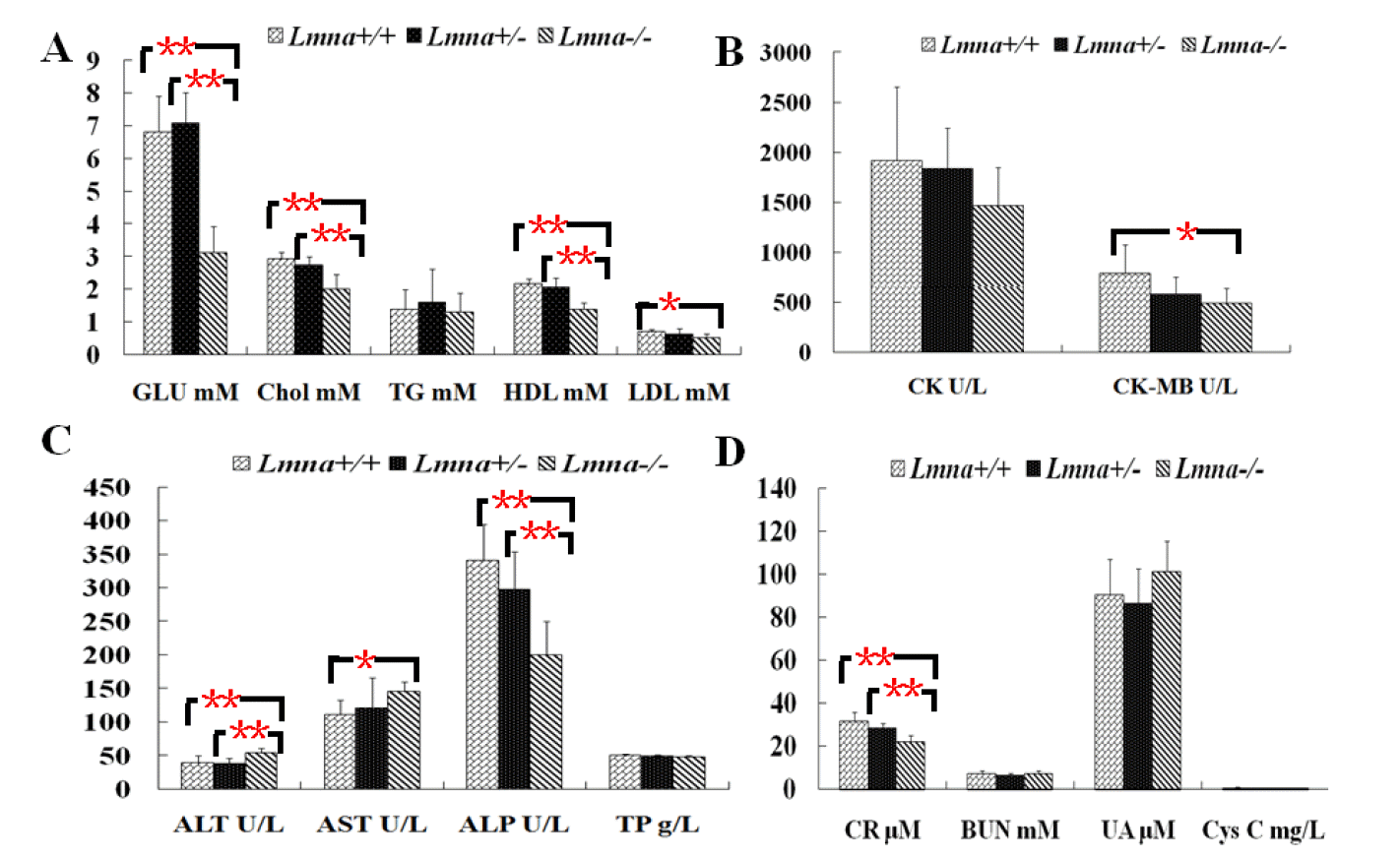
To explore the effect of Lmna deficiency on metabolism, we measured blood biochemical parameters of the mice on the 28th day after their birth. Ten serum samples from each group were taken for 15 analyses including glycolipid metabolism, heart function, liver function, and kidney function tests. It was found that the level of ALT (Table 1 and Figure 5C) in LmnaŌłÆ/ŌłÆ mice was significantly higher than that of other 2 groups (p<0.05). Compared with Lmna+/+ and Lmna+/ŌłÆ mice, LmnaŌłÆ/ŌłÆ mice exhibited lower levels of glucose, cholesterol, HDL (Figure 5A), ALP (Figure 5C) and creatinine (Figure 5D) (p<0.05). The AST levels in LmnaŌłÆ/ŌłÆ mice were higher than Lmna+/+ mice, not those of Lmna+/ŌłÆ mice, while LDL and CK-MB levels were significantly lower (Figure 5B) (p<0.05). There were no significant differences of triacylglycerols, creatine kinase, total protein, BUN, cystatin C and uric acidin Lmna+/+, Lmna+/ŌłÆ and LmnaŌłÆ/ŌłÆ mice (p>0.05). As expected, there were no significant difference of the 15 parameters in male and female mice (data not shown, p>0.05).
Newborn Lmna deficient mice were indistinguishable from heterozygous or wild-type mice. However, within 2 to 3 weeks, a reduction in their body weight was noticed, and this slower growth continued up to 4 weeks. Despite normal tooth development and the continued ability to eat, the growth of these mice had ceased. At week 4, their mean body weight was approximately 50% that of their wild-type or heterozygous littermates (Sullivan et al., 1999). Through genotyping, we also found that mouse growth was consistent with the growth rate reported by Sullivan (Sullivan et al., 1999). We found that the average body weights of Lmna+/+ and Lmna+/ŌłÆmice were larger than LmnaŌłÆ/ŌłÆmice approximately 19 days after birth, and after 28 days, the average weights of Lmna+/+ and Lmna+/ŌłÆ mice were approximately twice that of LmnaŌłÆ/ŌłÆ mice. We calculated and compared the weights of different organs from the three groups of mice and found that homozygous deletion of Lmna significantly decreased weights and increased the ratio of organ to body weight in most of tissues. These ratios increased in mutant mice may be due to organ functional increase or pathological enlargement. Combined with the biochemical indicators of different organs function and the former literature analysis (Wolf et al., 2008), kidney may be functional increased while the heart is pathological enlarged.
Because the mice that are homozygous for the Lmna null allele began to display abnormal gait with a stiff walking posture at 3 to 4 weeks and all had died by the eighth week, we performed serological analyses of all mice at the age of 4 weeks while the animals were still in good health and were mobile. This time point was selected due to the short life-span of the homozygous mice, and it enabled us to observe changes in serum biochemistry that accompanied externally apparent phenotypic changes in LmnaŌłÆ/ŌłÆ mice. The serological changes may indicate which tissue dysfunctions occurred and led to the early death of LmnaŌłÆ/ŌłÆ mice. The heterozygotes were also apparently normal and did not exhibit premature mortality compared with their wild-type siblings.
To the best of our knowledge, this is the first comprehensive report of 13 blood biochemical parameters except for glucose and triacylglyceride (Cutler et al., 2002) in LmnaŌłÆ/ŌłÆ and Lmna+/ŌłÆ C57BL/6 mice. Our results were consistent with two parameters reported (Cutler et al., 2002). Therefore, our data may aid in the evaluation of changes to metabolism in these experimental animals compared to control animals. Compared with previous reports, our data regarding wild-type C57BL/6 mice at 4 weeks of age demonstrated some similar results, while some parameters were higher or lower (Kubben et al., 2002; Schnell et al., 2002; Zhou and Hansson, 2004; Mazzaccara et al., 2008). The discrepancies in the biochemical values might depend on differential feeding of the mice, the ages of the mice at the time when blood was taken, the sex of the mice, or the environmental conditions in the laboratory in which the mice were reared (Yan et al., 2004; Zhou and Hansson, 2004; Mazzaccara et al., 2008). In addition to the above-mentioned factors, the site of blood collection may play a substantial role in the variation of both the biochemical values and the clinical pathology results (Schnell et al., 2002).
Although the numerical values of the total protein and triacylglyceride parameters of LmnaŌłÆ/ŌłÆ mice were less than those of other genotypes mice, the trend was not significantly different. Several other serum metabolic indicators that were measured showed lower levels in LmnaŌłÆ/ŌłÆ mice compared to Lmna+/+ mice, including glucose, cholesterol, HDL and LDL. Consistent with the observed weight reduction in LmnaŌłÆ/ŌłÆ mice, glucose levels were significantly reduced in these animals. Our results suggested that not only lipoproteins diminished but also the level of glucose decreased in the disorders observed in LmnaŌłÆ/ŌłÆ mice. These changes may be the result of reduced hepatic glucose production because we found that the pyruvate carboxylase (Pcx), the first of the key enzymes involved in glyconeogenesis, in LmnaŌłÆ/ŌłÆ mice was about 71.76% of the level in wild mice (data not shown). HDL is an independent cardiovascular risk factor and that an increase of HDL of only 10 mg/L leads to a risk reduction of 2% to 3% (Mahdy Ali et al., 2012). Therefore HDL reduction also could accelerate coronary heart disease considerably in Lmna-deleted mice.
The level of creatine kinase was 1,913┬▒737.4 U/L in Lmna+/+ mice and was the closest to the average value reported by Michael et al. (Schnell et al., 2002) in which creatine kinase activity was 2,114┬▒1,571 U/L. The similar results may be because the blood samples were collected via the same retro-orbital blood collection method, and this sampling method appears to have introduced inter-individual differences that skew the results. Therefore, we suggest that this method of blood sampling is not suitable for detection of kinase activity and it may also reduce the experimental significance of differences detected in mice of different genotypes. The isozyme CK-MB and AST value, two indices of cardiomyocyte damage, were significantly different between the LmnaŌłÆ/ŌłÆ and Lmna+/+ groups. The AST levels were higher than those of the Lmna+/+ group, which most likely indicates that the Lmna deletion leads to myocardial injury or liver alterations in mice. The lower CK-MB levels may be due to myocardial hypoplasia in LmnaŌłÆ/ŌłÆ mice.
ALT is a sensitive index that reflects liver injury. The ALT levels in LmnaŌłÆ/ŌłÆ mice were significantly higher than those of Lmna+/+ and Lmna+/ŌłÆ mice (p<0.05). Our results suggest that the liver organ function of 4-week-old LmnaŌłÆ/ŌłÆ mice may be affected by the degree of liver cell injury that is indicated by the increase in ALT level. Increased ALT levels may be caused by the damage of cardiac or skeletal muscle or other tissues. Liver damage can also lead to increased ALP, but in LmnaŌłÆ/ŌłÆ mice, ALP is reduced compared with the other two groups.
Creatinine, urea, uric acid and cystatin C serum analyses are used to evaluate kidney function (Rathkolb et al., 2009). Urea is the primary parameter used to evaluate renal function. Recent research demonstrates that there is a high correlation between impaired renal function and cystatin C concentrations in the serum, which can reflect changes in the human glomerular filtration rate (GFR). These tests are used as early diagnostic tools for diabetic nephropathy and hypertension-associated renal damage. The serum concentrations of urea, cystatin C and uric acid showed no significant differences among the three different Lmna genotypes in the mice from this study. However, creatinine is a less sensitive evaluation index for renal function and was obviously lower in LmnaŌłÆ/ŌłÆ mice than in Lmna+/+ and Lmna+/ŌłÆ mice, which may suggest that renal function has not been damaged in LmnaŌłÆ/ŌłÆ mice. The reduction in serum creatinine may be due to muscle hypoplasia in LmnaŌłÆ/ŌłÆ mice because creatinine is a metabolic end-product of muscle creatine and phosphocreatine consumption, and its production is proportional to muscle mass.
In conclusion, the deletion of the Lmna gene not only affects the growth and development of C57BL/6 mice but also altered their serological biochemistry. Homozygous deletions of the Lmna gene can lead to impaired heart and liver function, as well as glucose and lipid metabolism disorders.
ACKNOWLEDGEMENTS
The authors would especially like to thank Professor Collin Stewart for kindly providing Lmna+/ŌłÆ mice. This work was funded by grants from the National Natural Science Foundation of China (30871440, 81170327), the Guangdong Natural Science Foundation (84524023 01001450, 9252402301000002), and the Science and Technology Innovation Fund of Guangdong Medical College (No. STIF201102, 1057113035, ZZDC009).
Figure┬Ā1
Genotyping of Lmna in the mice. PCR was applied to detect the Lmna allele in the mice. The upper bands indicate wild-type alleles. The lower bands indicate mice that possess the Lmna homozygous deletion. Mice with both bands are heterozygous. M = DL2000 Marker. 1ŌĆō10 = PCR amplification products from mice No. 1 to No. 10. P = Positive control. N = Negative control.
Figure┬Ā2
Growth curves of mice with different Lmna genotypes. The body weights of LmnaŌłÆ/ŌłÆ, Lmna+/+ and Lmna+/ŌłÆ mice were evaluated once every 3 days from their birth (D1) to 4 weeks (D28). The data are presented as the mean┬▒SD, n = 30. The black solid line denotes wild-type mice. The long dashed line denotes Lmna+/ŌłÆ mice. The short dashed line indicates LmnaŌłÆ/ŌłÆ mice. * p<0.05.
Figure┬Ā3
Tissue weights of Lmna+/+, Lmna+/ŌłÆ and LmnaŌłÆ/ŌłÆ mice. Most tissue weights of Lmna-deleted mice at 4 weeks after birth were decreased. The data are presented as the mean┬▒SD, n = 30. ** Indicates p<0.01.
Figure┬Ā4
The ratio of organ to body weight in Lmna+/+, Lmna+/ŌłÆ and LmnaŌłÆ/ŌłÆ mice. LmnaŌłÆ/ŌłÆ mice at 4 weeks displayed increased ratio of organ to body weight in most of tissues. The data are presented as the mean┬▒SD, n = 30. * Indicates p<0.05; ** Indicates p<0.01; *** Indicates p<0.001.
Figure┬Ā5
Blood biochemical values of Lmna+/+, Lmna+/ŌłÆ and LmnaŌłÆ/ŌłÆ mice. (A) Glycolipid metabolism correlation index; (B) Cardiac function indicators; (C) Liver function indices; (D) Renal function indices. The data are presented as the mean┬▒SD, n = 30. * Indicates p<0.05; ** Indicates p<0.001.
Table┬Ā1
Serum biochemical analytes (mean┬▒SD) measured in C57BL/6 mice with the genotypes Lmna+/+, Lmna+/ŌłÆ and LmnaŌłÆ/ŌłÆ
| Biochemical indexes | Lmna+/+ | Lmna+/ŌłÆ | LmnaŌłÆ/ŌłÆ | P(SNK) | ||
|---|---|---|---|---|---|---|
|
|
||||||
| n1 = 10 | n2 = 10 | n3 = 10 | n1n3 | n2n3 | n1n2 | |
| GLU (mmol/L) | 6.82┬▒1.09 | 7.09┬▒0.93 | 3.13┬▒0.78 | 0.000** | 0.000** | 1.000 |
| Chol1 (mmol/L) | 2.92┬▒0.21 | 2.74┬▒0.26 | 2.02┬▒0.42 | 0.000** | 0.000** | 0.560 |
| TG (mmol/L) | 1.38┬▒0.61 | 1.61┬▒1.01 | 1.3┬▒0.57 | 1.000 | 1.000 | 1.000 |
| HDL (mmol/L) | 2.17┬▒0.15 | 2.07┬▒0.26 | 1.38┬▒0.21 | 0.000** | 0.000** | 0.907 |
| LDL (mmol/L) | 0.7┬▒0.07 | 0.63┬▒0.16 | 0.53┬▒0.10 | 0.010* | 0.173 | 0.700 |
| CK (U/L) | 1,913┬▒737.4 | 1,838.2┬▒406.8 | 1,461.9┬▒383.88 | 0.210 | 0.381 | 1.000 |
| CK-MB (U/L) | 784.6┬▒286.8 | 529.7┬▒105.34 | 489.4┬▒149.48 | 0.013* | 1.000 | 0.117 |
| ALT (U/L) | 39.2┬▒10.21 | 37.6┬▒8.3 | 54.1┬▒6.4 | 0.002** | 0.001** | 1.000 |
| AST (U/L) | 110.4┬▒21.86 | 120.8┬▒44.6 | 145.4┬▒14.37 | 0.043* | 0.229 | 1.000 |
| ALP (U/L) | 340.6┬▒53.85 | 297┬▒56.62 | 200.1┬▒49.98 | 0.000** | 0.001** | 0.239 |
| TP (g/L) | 50.12┬▒2.03 | 48.63┬▒2.23 | 47.86┬▒2.22 | 0.081 | 1.000 | 0.406 |
| CR (╬╝mol/L) | 31.9┬▒3.96 | 28. 6┬▒2.32 | 22.2┬▒3.05 | 0.000** | 0.000** | 0.084 |
| BUN (mmol/L) | 7.26┬▒1.27 | 6.74┬▒0.89 | 7.3┬▒1.22 | 1.000 | 0.866 | 0.973 |
| UA (╬╝mol/L) | 90.6┬▒16.65 | 86.5┬▒16.19 | 101.4┬▒14.16 | 0.407 | 0.130 | 1.000 |
| Cys C (mg/L) | 0.77┬▒0.17 | 0.66┬▒0.09 | 0.64┬▒0.2 | 0.244 | 1.000 | 0.381 |
REFERENCES
Andres V, Gonzalez JM. 2009. Role of A-type lamins in signaling, transcription, and chromatin organization. J Cell Biol 187:945ŌĆō957.



Arancio W. 2012. A bioinformatics analysis of Lamin-A regulatory network: a perspective on epigenetic involvement in Hutchinson-Gilford progeria syndrome. Rejuvenation Res 15:123ŌĆō127.


Choi Bong Hwan, Lee JS, Lee SH, Kim SC, Kim SW, Kim KS, Lee JH, Seong HH, Kim TH. 2012. Porcine LMNA is a positional candidate gene associated with growth and fat deposition. Asian-Aust J Anim Sci 25:1649ŌĆō1659.



Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau M, Toniolo D, Schwartz K. 1999. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet 21:285ŌĆō288.


Cao H, Hegele RA. 2000. Nuclear lamin A/C R482Q mutation in canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum Mol Genet 9:109ŌĆō112.


Cutler DA, Sullivan T, Marcus-Samuels B, Stewart CL, Reitman ML. 2002. Characterization of adiposity and metabolism in Lmna-deficient mice. Biochem Biophys Res Commun 291:522ŌĆō527.


De Sandre-Giovannoli A, Chaouch M, Kozlov S, Vallat JM, Tazir M, Kassouri N, Szepetowski P, Hammadouche T, Vandenberghe A, Stewart CL, Grid D, Levy N. 2002. Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse. Am J Hum Genet 70:726ŌĆō736.



Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. 2003. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423:293ŌĆō298.



Jahn D, Schramm S, Schnolzer M, Heilmann CJ, de Koster CG, Schutz W, Benavente R, Alsheimer M. 2012. A truncated lamin A in the Lmna (ŌłÆ/ŌłÆ) mouse line: Implications for the understanding of laminopathies. Nucleus 3:463ŌĆō474.



Kubben N, Voncken JW, Konings G, van Weeghel M, van den Hoogenhof MM, Gijbels M, van Erk A, Schoonderwoerd K, van den Bosch B, Dahlmans V, Calis C, Houten SM, Misteli T, Pinto YM. 2011. Post-natal myogenic and adipogenic developmental: defects and metabolic impairment upon loss of A-type lamins. Nucleus 2:195ŌĆō207.



Mahdy Ali K, Wonnerth A, Huber K, Wojta J. 2012. Cardiovascular disease risk reduction by raising HDL cholesterol - current therapies and future opportunities. Br J Pharmacol 167:1177ŌĆō1194.



Mazzaccara C, Labruna G, Cito G, Scarfo M, De Felice M, Pastore L, Sacchetti L. 2008. Age-related reference intervals of the main biochemical and hematological parameters in C57BL/6J, 129SV/EV and C3H/HeJ mouse strains. PLoS One 3:e3772



Mendez-Lopez I, Worman HJ. 2012. Inner nuclear membrane proteins: impact on human disease. Chromosoma 121:153ŌĆō167.


Muchir A, van Engelen BG, Lammens M, Mislow JM, McNally E, Schwartz K, Bonne G. 2003. Nuclear envelope alterations in fibroblasts from LGMD1B patients carrying nonsense Y259X heterozygous or homozygous mutation in lamin A/C gene. Exp Cell Res 291:352ŌĆō362.


Raffaele Di, Barletta M, Ricci E, Galluzzi G, Tonali P, Mora M, Morandi L, Romorini A, Voit T, Orstavik KH, Merlini L, Trevisan C, Biancalana V, Housmanowa-Petrusewicz I, Bione S, Ricotti R, Schwartz K, Bonne G, Toniolo D. 2000. Different mutations in the LMNA gene cause autosomal dominant and autosomal recessive Emery-Dreifuss muscular dystrophy. Am J Hum Genet 66:1407ŌĆō1412.



Rathkolb B, Noyes HA, Brass A, Dark P, Fuchs H, Gailus-Durner V, Gibson J, de Angelis MH, Ogugo M, Iraqi F, Kemp SJ, Naessens J, Pope ME, Wolf E, Agaba M. 2009. Clinical chemistry of congenic mice with quantitative trait loci for predicted responses to Trypanosoma congolense infection. Infect Immun 77:3948ŌĆō3957.



Schnell MA, Hardy C, Hawley M, Propert KJ, Wilson JM. 2002. Effect of blood collection technique in mice on clinical pathology parameters. Hum Gene Ther 13:155ŌĆō161.


Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol 147:913ŌĆō920.



Wang JP, Yan L, Lee JH, Kim IH. 2013. Evaluation of bacteriophage supplementation on growth performance, blood characteristics, relative organ weight, breast muscle characteristics and excreta microbial shedding in broilers. Asian-Aust J Anim Sci 26:573ŌĆō578.




Wolf CM, Wang L, Alcalai R, Pizard A, Burgon PG, Ahmad F, Sherwood M, Branco DM, Wakimoto H, Fishman GI, See V, Stewart CL, Conner DA, Berul CI, Seidman CE, Seidman JG. 2008. Lamin A/C haploinsufficiency causes dilated cardiomyopathy and apoptosis-triggered cardiac conduction system disease. J Mol Cell Cardiol 44:293ŌĆō303.



Worman HJ, Ostlund C, Wang Y. 2010. Diseases of the nuclear envelope. Cold Spring Harb. Perspect. Biol. 2:a000760



- TOOLS





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





