Gene Expression Profiling in the Pituitary Gland of Laying Period and Ceased Period Huoyan Geese
Article information
Abstract
Huoyan goose is a Chinese local breed famous for its higher laying performance, but the problems of variety degeneration have emerged recently, especially a decrease in the number of eggs laid. In order to better understand the molecular mechanism that underlies egg laying in Huoyan geese, gene profiles in the pituitary gland of Huoyan geese taken during the laying period and ceased period were investigated using the suppression subtractive hybridization (SSH) method. Total RNA was extracted from pituitary glands of ceased period and laying period geese. The cDNA in the pituitary glands of ceased geese was subtracted from the cDNA in the pituitary glands of laying geese (forward subtraction); the reverse subtraction was also performed. After sequencing and annotation, a total of 30 and 24 up and down-regulated genes were obtained from the forward and reverse SSH libraries, respectively. These genes mostly related to biosynthetic process, cellular nitrogen compound metabolic process, transport, cell differentiation, cellular protein modification process, signal transduction, small molecule metabolic process. Furthermore, eleven genes were selected for further analyses by quantitative real-time PCR (qRT-PCR). The qRT-PCR results for the most part were consistent with the SSH results. Among these genes, Synaptotagmin-1 (SYT1) and Stathmin-2 (STMN2) were substantially over-expressed in laying period compared to ceased period. These results could serve as an important reference for elucidating the molecular mechanism of higher laying performance in Huoyan geese.
INTRODUCTION
The Huoyan goose was listed as one of the seventy-eights nationally protected domestic animals by the Chinese government in the year of 2000. It was used for both meat and egg production, and was famous for its higher laying performance. But breed selection and conservation work for this famous Chinese local breed is relatively weak; the problems of variety degeneration have been very prominent, especially the number of laying eggs decreased.
In poultry, the reproductive endocrine system and the reproductive activity are strictly controlled by the hypothalamic-pituitary-gonadal axis (Padmanabhan et al., 2002). The laying performance is determined by the number of follicles destined for ovulation and the capacity of the oviduct to transform the ova into a hardshelled egg. The egg-production process, from follicle maturation to ovulation, is strictly controlled by hormones and other physiological conditions, including energy metabolism and the regulating of apoptosis of oocyte and follicle-associated cells. The activation and maintenance of normal follicular functions are dependent on gonadotropins including luterinizing hormone (LH) and prolactin (PRL) secreted by the pituitary, which in turn is regulated by the hypothalamic gonadotropin releasing hormones (GnRH) (Kuo et al., 2005). Minute differences in hypothalamic or pituitary functions, might affect reproductive activities, including folliculogenesis, ovulation, oviposition, and incubation behavior (Etches et al., 1984).
The goose is a seasonal breeder (laying during the short photoperiodic season) with a low egg production rate. The pituitary LH and PRL together with other hormones, function to synchronize or regulate the reproductive seasonality in both long and short day breeding birds (Shi et al., 2007). Consequently, the pituitary gland is ideal tissues to elucidate the molecular mechanism associated with egg production, since genes may be transcribed differently under different physiological conditions. Information of gene profiles in pituitary gland at different reproduction period is essential for studying the molecular genetic mechanisms of egg laying and their application to improving the laying performance.
Suppression subtractive hybridization (SSH) is a method to get a gene expression profile specifically designed for comparing gene expression at various developmental stages or in altered physiological conditions (Diatchenko et al., 1996). This powerful technique enables comparing mRNA of two populations and then obtaining clones of genes that are differentially expressed in one population but not in another. First, both mRNA populations are converted into cDNA, which the cDNA that contains differentially expressed transcripts as tester, and the reference cDNA as driver. Tester and driver cDNAs are hybridized and then the hybrid common sequences are removed. Consequently, the remaining unhybridized cDNAs represent genes that are differentially expressed in the tester yet absent from the driver mRNA. This allows isolation of genes that are specifically expressed under diverse sets of conditions. Based on this technology, nine transcripts related to high egg production in the chicken hypothalamus/pituitary gland were identified (Shiue et al., 2006). As for the goose, 18 known and 8 unknown differentially expressed genes in the ovaries of egg-laying and pre-laying Zi geese were identified by this method (Kang et al., 2009). In addition, it was also used to detect differential expression of genes in the livers of laying and pre-laying geese (Ding et al., 2007). Although the differential expression profile of genes in the pituitary gland has been studied in laying geese compared with pre-laying geese (Yen et al., 2006), but to the best of our knowledge, information of Huoyan geese in laying period and ceased period has not been investigated so far.
In this study, gene expression profiles in the pituitary gland of laying period and ceased period Huoyan geese were investigated using SSH coupled with qRT-PCR methods. Combined results will be useful in elucidating the molecular mechanisms that underlie egg laying and improving the laying performance of Huoyan geese.
MATERIALS AND METHODS
Geese and tissue collection
The animal protocol used in this experiment was approved by the Animal Welfare Committee of Liaoning Province, China. The Huoyan geese were selected from Panjin Jiyuan goose breeding farm and raised according to the program used at this farm. During the experiment, geese were fed ad libitum with rice grain and were supplemented with green grass or water plants whenever possible. Feed was given during the daytime when the geese were released into an open area outside the house. Twenty female geese were killed by exsanguinations in January to obtain pituitary samples of ceased period geese. Another twenty female geese were killed in June to obtain pituitary samples of laying period geese. All pituitary samples were quickly dissected, frozen in liquid nitrogen, and stored at −80°C until total RNA extraction.
Total RNA isolation and reverse transcription
Total RNA was prepared by TRIzol reagent (Invitrogen Corporation, Carlsbad, CA, USA) according to the instruction of the manufacturer. The total RNA samples from ceased period and laying period pituitary (n = 20, for each) were pooled separately. The first-strand cDNA and ds-cDNA were synthesized using SMARTer™ PCR cDNA Synthesis Kit (Clontech, Mountain View, CA, USA) and later, the ds-cDNA was purified with QIAquick PCR Purification Kit (Qiagen, Hilden, Germany).
Construction of the suppression subtractive hybridization cDNA library
The cDNA libraries were constructed by SSH using a PCR-Select™ cDNA Subtraction Kit (Clontech) following manufacturer protocol. A forward SSH library was constructed to isolate the up-regulated genes of the laying period. It was used to identify clones in which the laying period cDNA was used as the tester and the ceased period cDNA as the driver. In contrast, a reverse SSH library was constructed to detect the down-regulated genes of the laying period. It was used to identify clones in which the ceased period cDNA was used as the tester and the laying period cDNA as the driver. After digestion with RsaI, the tester cDNA was divided into two subpopulations, and then each of which was ligated to a unique adaptor at 16°C for 8 h. The adaptor 1-ligated or adaptor 2R-ligated tester cDNA was separately hybridized at 68°C for 8 h with an excess of driver cDNA after denaturation at 98°C for 90 s. The two hybridized samples were then mixed together without denaturation and hybridized once more at 68°C for 12 h with excess of denatured driver cDNA. The resulting mixture was added with 200 μl dilution buffer and amplified by two rounds of suppression PCR. The primary PCR was performed using PCR primers in the kit. After the reaction mixtures were incubated at 75°C for 5 min to extend the adaptors, PCR was performed at 94°C for 10 s, 66°C for 30 s, and 72°C for 90 s for 27 cycles in a reaction volume of 25 μl. The PCR product was then diluted to 10-fold, from which 10 diluted liquid was used as template in the subsequently nested PCR. This time, the reaction was performed for 12 cycles of 94°C for 10 s, 68°C for 30 s, and 72°C for 90 s, using nested PCR primer1 and primer 2R. The PCR products obtained from the forward and reverse subtractions were separately cloned into the pMD-18-T vector (TaKaRa, Dalian, China), and transformed into Escherichia coli DH-5α to establish different cDNA libraries.
DNA sequencing and bioinformatics analysis
Positive clones were selected by conventional blue-white screening. White clones were randomly selected from both forward and reverse libraries. The positive clones were further validated by colony PCR using nested PCR primers provided in the kit. The resulting products were subjected to the direct sequencing with M13 primers. The vector sequences were removed through a Perl script and checked through VecScreen (http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html). Sequence analysis and homology comparisons were carried out using the BLASTn program of NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Gene Ontology (GO) annotations were performed online from GORetriever (http://www.agbase.msstate.edu/). If no annotation was returned, GOanna was used to retrieve GO annotations assigned on the basis of sequence similarities. The resulting annotations were summarized according to the generic GOSlim set using GOSlimViewer (McCarthy et al., 2007).
Quantitative RT-PCR
To validate the differentially expressed genes detected by the SSH approach, eleven ESTs (expressed sequence tags) which six from forward library and five from reverse library were selected for qRT-PCR analysis. The qRT-PCR primers were designed using Primer 3.0 (http://frodo.wi.mit.edu/primer3). All the information of the primers is listed in Table 1. Total RNA was extracted using TRIzol Reagent (Invitrogen Corporation, Carlsbad, CA, USA) according to the manufacturer’s instructions. The concentration and purity of the RNA were measured using a spectrophotometer. Two micrograms of total RNA was reverse transcribed using PrimerScript RT reagent Kit (TaKaRa, Dalian, China). Real-time PCR was carried out on LightCycler® 480 II real-time PCR system (Roche). Each 25 μl reaction volume contained 1 μl 10 μM (each) forward and reverse primers, 12.5 μl 2×SYBR® Premix Ex Taq™ II (Takara, Dalian, China), and 2 μl cDNA products, and the final volume was adjusted using PCR-water. The following PCR program was used for amplification: 15 min at 95°C, 40 cycles of denaturation at 95°C for 10 s and annealing and extension at 60°C for 30 s. Relative gene expression was calculated using the 2−ΔΔCt method, using the ceased period group as calibrator (relative expression = 1) and β-actin as an internal reference gene.
Statistical analysis was carried out by student’s t tests with SPSS 16.0 for Windows (SPSS Inc.) to evaluate whether the means were significantly different (p<0.05).
RESULTS
Differential screening and EST sequencing
Based on the results of the differential screening, 112 cDNA clones from the forward library and 110 cDNA clones from the reverse library were randomly picked and sequenced. After trimming, 103 and 104 high quality ESTs from the forward and reverse library, respectively, were obtained, all the processed ESTs were submitted to NCBI for BLASTn.
Analysis of BLAST results revealed that in forward library, as shown in Table 2, 30 sequences have high degree of similarity with known or predicted genes in GenBank, such as P21 protein (Cdc42/Rac)-activated kinase 2, transcript variant 2 (PAK2), Stathmin-2 (STMN2), Synaptotagmin-1(SYT1), Sprouty-related, EVH1 domain containing 1 (SPRED1), Protocadherin 9 (PCDH9) and Vesicle-associated membrane protein-associated protein A (VAPA). In reverse library, as shown in Table 3, 24 sequences have high degree of similarity with genes like Vesicle-associated membrane protein 4 (VAMP4), Calmodulin binding transcription activator 1 (CAMTA1), SPARC related modular calcium binding 2 (SMOC2), Cyclin-dependent kinase 5, regulatory subunit 1 (CDK5R1) and Ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENPP2).
Functional annotation
Based on the gene ontology (GO) terms, these genes were functionally annotated. As illustrated in Figure 1, majority of the biological processes (such as anatomical structure development, biosynthetic process, cellular nitrogen compound metabolic process, transport, cell differentiation, cellular protein modification process, signal transduction, small molecule metabolic process, anatomical structure formation involved in morphogenesis, transmembrane transport, cell death, translation, and DNA metabolic process), some cellular components (such as cell, intracellular, organelle, cytoplasm, mitochondrion, nucleus, protein complex and plasma membrane), and some molecular functions (such as kinase activity, ion binding and enzyme regulator activity) were presented in both libraries. Other GO terms, such as transmembrane transport, ribosome biogenesis, growth, endosome, nucleolus, ligase activity, lyase activity, lipid binding, peptidase activity and methyltransferase activity were identified only in the forward library. Vice versa, some GO terms were identified only in the reverse library, such as cytoskeleton organization, cell motility, lipid metabolic process, cell morphogenesis, cell cycle, protein targeting, membrane organization, neurological system process, locomotion, developmental maturation, vesicle-mediated transport, embryo development, cell proliferation, cell-cell signaling, plasma membrane organization, immune system process, proteinaceous extracellular matrix, cytosol, Golgi apparatus, endoplasmic reticulum, extracellular region, transferase activity, transferring acyl groups, nucleic acid binding transcription factor activity and cytoskeletal protein binding.
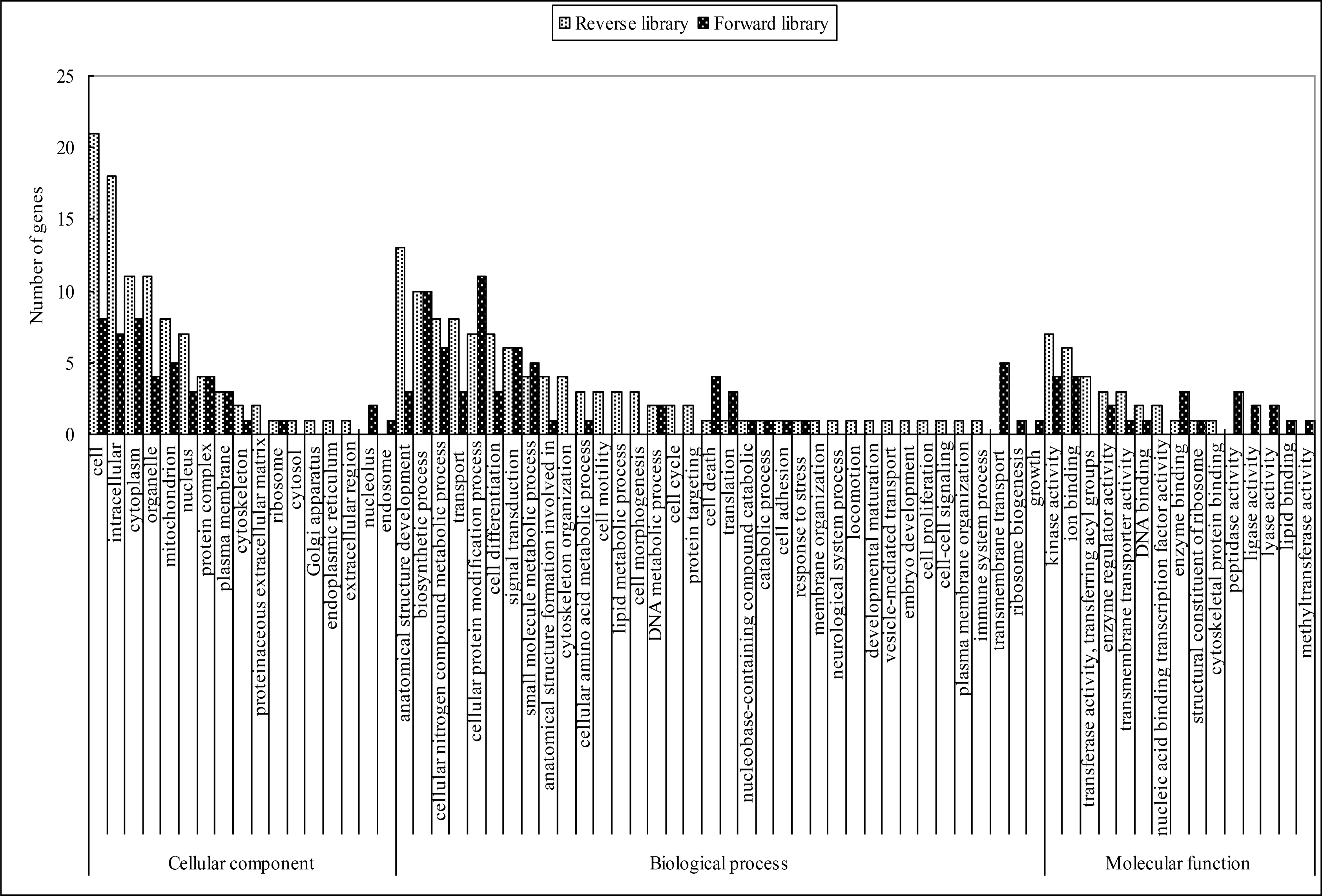
Gene ontology (GO) classification of the differentially expressed ESTs in the forward and reverse SSH libraries. Based on the GO terms, the ESTs are categorized into putative functional groups. The black and white columns represent the up- and down-regulated genes from forward and reverse libraries, respectively.
Confirmation of gene expression with quantitative RT-PCR
To verify the results of SSH, we analyzed the expression of 11 ESTs by qRT-PCR. The fold difference between the laying group and the ceased group was calculated by 2−ΔΔCt method. As illustrated in Figure 2, the qRT-PCR results for the most part, consistent with the SSH results. For the 6 up-regulated genes, 4 of them were over-expressed in the laying period, whereas 4 out of 5 down-regulated genes were under-expressed. Particularly, SYT1, STMN2, ENPP2, and CDK5R1 exhibited significantly differentially expressed in laying period pituitary compared with that in ceased period pituitary.
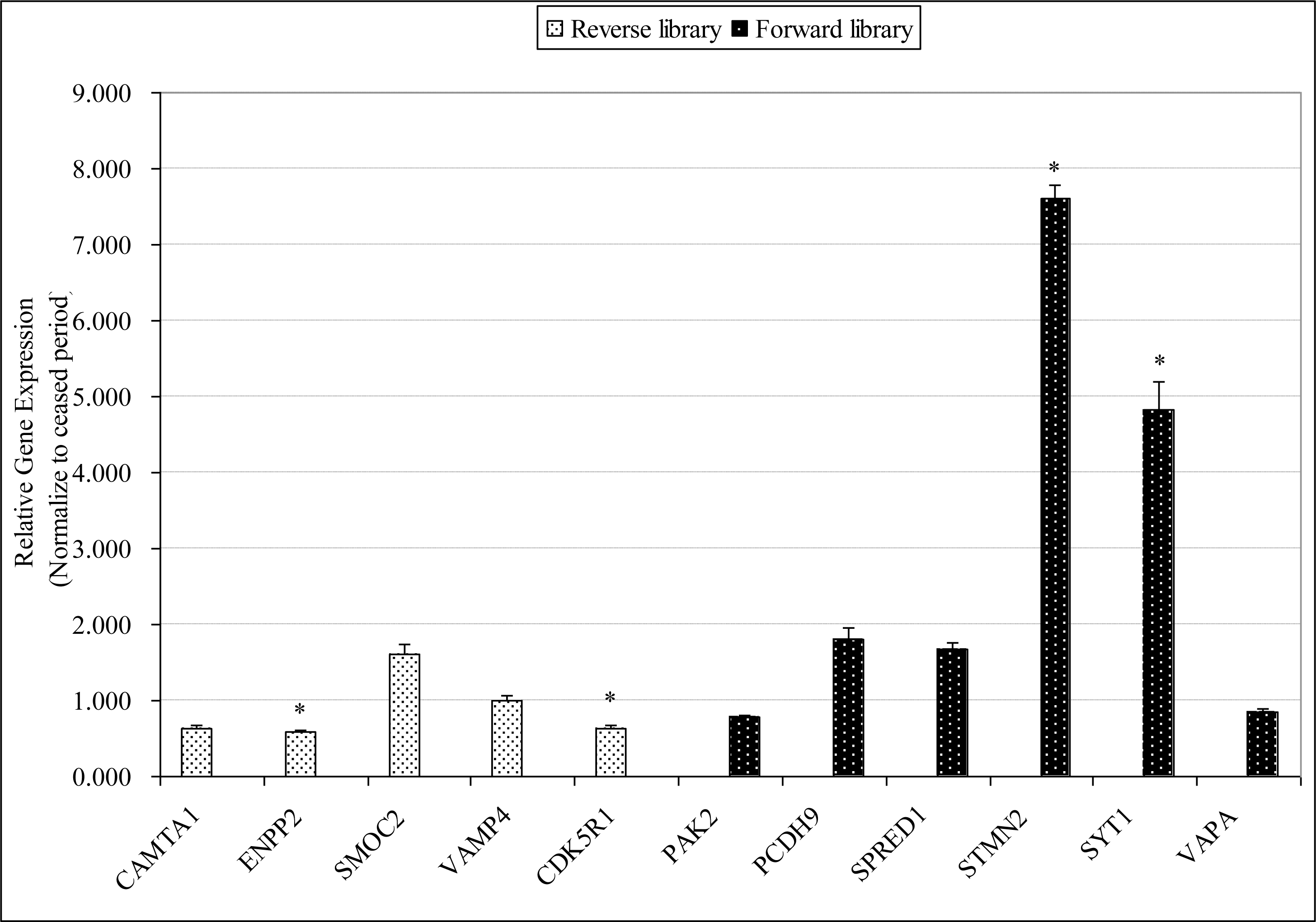
Quantitative real-time PCR validation. The gene expression level of 11 randomly selected ESTs, including 6 from SSH forward library (the black column) and 5 from SSH reverse library (the white column), was tested using qRT-PCR. The relative gene expression level in the laying period was normalized to the ceased period. Data are presented as mean±SE. Asterisks denote significant gene expression differences between the laying period and ceased period, as determined by the student’s t tests (* p<0.05).
DISCUSSION
In the present work, we employed SSH method to identify differentially expressed genes in the pituitary gland of Huoyan geese between laying period and ceased period. Some genes involved in signal transduction (such as CNTNAP2, STMN2, PAK2, and SMOC2), calcium ion binding (such as CAMTA1, PCDH9, CDK5R1, and SYT1), MAPK signaling pathways (SPRED1), I-kappaB kinase/NF-kappaB signaling pathways (VAPA), and transport (such as VAMP4, SLC5A11, SLC4A4, COX1, and COX3) were identified. Subsequently, eleven of those including PAK2, PCDH9, SPRED1, STMN2, SYT1, VAPA, CAMTA1, ENPP2, SMOC2, VAMP4, and CDK5R1 were selected for further validating using qRT-PCR. The results were mostly consistent with the SSH results. Interestingly, several of these genes might be involved secretion of hormones.
One of the major findings was that several genes related to exocytosis were found differentially expressed in our study. Exocytosis is a key biological process that controls the neurotransmission and release of secretory products from neurones and other secretory cell types. Neurotransmitters, hormones, or other secretory products are packed in vesicles, a number of these vesicles fuse with the surface membrane, during both nonactivated and activated phases, to release secretory product into the extracellular space (Knight and Baker, 1987). Neurotransmitters and hormones release is triggered by Ca2+ binding to a presynaptic Ca2+ sensor that induces synaptic vesicle exocytosis with a high degree of Ca2+ cooperativity. Synaptotagmin-1 (SYT1) has been identified as the primary Ca2+ sensors for synaptic vesicle exocytosis and a major transducer of Ca2+ signaling in membrane fusion events and regulated secretion (Koh and Bellen, 2003; Kreft et al., 2003; Shin et al., 2009). There is evidence that SYT1 plays a physiological role in secretion by differentiated pituitary cells. Genetic experiments in mice have demonstrated that SYT1 mutants have defects in regulated secretion (Koh and Bellen, 2003). Mice with homozygous disruption of the SYT1 die shortly after birth and have defects in neurotransmitter release from hippocampal neurons (Geppert et al., 1994). The pituitary-specific transcription factor (POU1F1) is a factor that bound to and activated the PRL and GH promoters, and is also important for proper development of the pituitary cells that express PRL, GH, and TSH. Study provides evidence that it can bind to a specific site in the SYT1 gene and that this binding contributes to activation of SYT1 expression. It seems likely that activation of SYT1 gene expression is part of a mechanism mediating POU1F-induced differentiation of pituitary cells and presumably contributes to the endocrine/secretory phenotype (Howard et al., 2009). Production and secretion of hormones by the pituitary involve highly orchestrated intracellular transport and sorting steps. Peptide hormones destined for regulated secretion are packaged into secretory granules that bud from the trans-Golgi network (TGN). VAMP4 (Vesicle-associated membrane protein 4), a member of VAMP proteins implicated in vesicle docking with membranes, has been suggested to play a role in mediating trans-Golgi network (TGN) trafficking. VAMP4 marks a novel vesicular sorting pathway during granule membrane remodeling, and Synaptotagmin IV follows the trafficking patterns of VAMP4 and modulates Ca2+-triggered exocytosis during granule maturation (Steegmaier et al., 1999).
It is well known that Ca2+ is the primary intracellular signaling molecule controlling the fusion of secretory vesicles with the plasma membrane to release transmitters from neurons and hormones from endocrine cells (Martin, 2003). Increase in intracellular Ca2+ concentration is a key signal to trigger exocytosis in most neuronal and endocrine tissues. Pituitary hormone secretion is a Ca2+-dependent process. Depriving extracellular Ca2+ by incubation in media containing Ca2+ buffers such as EGTA inhibits GnRH-induced LH secretion (BATES and CONN, 1984), GRF-induced GH secretion (Sheppard et al., 1980) and TRH-induced PRL secretion (Thorner et al., 1980). Furthermore, this inhibition can be reversed by elevation of extracellular calcium. In this study, except SYT1, several other genes which involved calcium ion binding were identified differentially expressed. Protocadherins (PCDHs) constitute the largest subgroup within the cadherin family of calcium-dependent cell-cell adhesion molecules, and have been suggested to play role in the formation and maintenance of the synaptic connections (Takeichi and Abe, 2005). Study demonstrated some PCDHs were involved in regulation of LHRH neuronal connections (Varju et al., 2009). Calmodulin can trigger exocytosis of already primed vesicles in endocrine cells; it also binds to the Ca2+ sensor synaptotagmin, and could be involved in synaptotagmin-related events (Langley and Grant, 1997). Calmodulin binding transcription activator 1 (CAMTA1) was differentially expressed in our study, it may be involved alterations of intracellular Ca2+ concentration and play a major role in anterior pituitary cell stimulus-secretion coupling.
Remarkably, the expression of Stathmin-2 (STMN2) was identified significantly up-regulated in laying period. Stathmin is the founding member of a family of proteins that play critically important roles in the regulation of the microtubule cytoskeleton, its expression and phosphorylation are regulated throughout development and in response to extracellular signals regulating cell proliferation, differentiation and functions (Rubin and Atweh, 2004). It is highly expressed in the nervous and neuroendocrine systems and its phosphorylation could be related to regulation of cell growth arrest, terminal differentiation and hormone secretion (Rowlands et al., 1995; Drouva et al., 1998). Studies demonstrated that Stathmin play a potential role in the regulation of hormone secretion in rodent pituitary and insulinoma cell lines (Rubin and Atweh, 2004). Interestingly, the mRNA expression level of STMN2 was found to be significantly higher in the hypothalamus and pituitary gland of high egg production chicken strains (Shiue et al., 2006). Its role in regulation of laying egg needs to be investigated in the future.
Furthermore, several genes involved in regulation of kinase activity were identified differentially expressed in this study. Sprouty-related proteins (SPREDs) with EVH1 (enabled/vasodilator-stimulated phosphoprotein homology 1) domain are membrane-associated suppressors of MAPK signaling, a major regulator of cellular proliferation and differentiation. SPRED2 was identified as a negative regulator of the Hypothalamic-Pituitary-Adrenal Axis, SPRED2 deficiency leads to an increased MAPK signaling, which results in an augmented its secretagogue corticotrophin releasing hormone (CRH) promoter activity, the subsequent CRH overproduction causes an up-regulation of downstream hypothalamic-pituitary-adrenal (HPA) hormone secretion (Ullrich et al., 2011). SPRED1 has been shown to be expressed predominantly in the brain and enriched in the central nervous system, where it mediates cortical development, neural stem cell proliferation, and vesicular trafficking (Phoenix and Temple, 2010). In this study, the differential expression of SPRED1 may be associated with its vesicular trafficking and negative regulation of the Ras-MAPK-ERK pathway during production and secretion of hormones in the pituitary gland. CDK5R1, also named p35 was the first neuronal-specific regulatory subunit of CDK5 (Cyclin-dependent kinase) identified via its physical interaction with CDK5, activating CDK5 after direct binding (Ko et al., 2001). Study suggest that CDK5/p35 play critical roles in neural migration, differentiation, and synaptogenesis in immature neurons. Cdk5/p35 also was indicated inhibits neurotransmitter release through the phosphorylation of P/Q-type voltage-dependent Ca2+ channel and down-regulation of the channel activity (Tomizawa et al., 2002). The brain neurotransmitters involve the mechanism triggering the release of the hypothalamic hormones, Changes in neurotransmitter output and in particular alterations in secretion of the monoamines, dopamine, noradrenaline and serotonin, have been associated with many hormonal changes in mammals (Chen et al., 1974; Advis et al., 1978). The effects of neurotransmitters on release of pituitary hormones in birds have been studied. Monoamines were confirmed to be capable of affecting pituitary release of PRL and GH in a pituitary-hypothalamus in pigeon (Hall, 1982).
In summary, we have shown gene expression profiles in the pituitary gland of laying period and ceased period Huoyan geese. Particularly, SYT1 and STMN2 which play an important role in release of pituitary hormones were first to demonstrate substantially over-expressed in the laying goose pituitary gland. Further demonstration of the functions of genes will add great value to the understanding of the reproductive biology of the goose.
Acknowledgements
We would like to thank the staff of Panjin Jiyuan goose breeding farm, who assisted in the collection of goose pituitary gland samples.
This study was supported by the National Natural Science Foundation of China (Grant No. 31172286).


