 |
 |
Abstract
The objective of this study was to investigate single nucleotide polymorphisms (SNPs) in the bovine nephroblastoma overexpressed (NOV) gene and to evaluate whether these polymorphisms affect carcass traits in the Korean cattle population. We resequenced to detect SNPs from 24 unrelated individuals and identified 19 SNPs within the full 8.4-kb gene, including the 1.5-kb promoter region. Of these 19 SNPs, four were selected for genotyping based on linkage disequilibrium (LD). We genotyped 429 steers to assess the associations of these four SNPs with carcass traits. Statistical analysis revealed that g.7801T>C and g.8379A>C polymorphisms in the NOV gene were associated with carcass weight (p = 0.012 and 0.008, respectively), and the g.2005A>G polymorphism was associated with the back fat thickness (BF) trait (p = 0.0001). One haplotype of the four SNPs (GGTA) was significantly associated with BF (p = 0.0005). Our findings suggest that polymorphisms in the NOV gene may be among the important genetic factors affecting carcass yield in beef cattle.
Carcass weight and back fat thickness are economically important cattle characteristics in the Korean beef industry. The retail carcass price reflects the sum of the quality and yield grades, and back fat thickness negatively affects the yield grade (Moon et al., 2003). Two factors that affect the growth rate of cattle are genetic potential, which regulates a complex of hormone and growth factors and their associated interactions, and environmental conditions such as nutrition, climate, disease and management (Hermesmeyer et al., 2000). Genetic potential has long been considered to be an important factor in the competitiveness of beef cattle production.
The bovine nephroblastoma overexpressed (NOV) gene is positioned near RM192 on bovine chromosome 14 (Lee et al., 2011). This region has also been reported to harbor a quantitative trait locus (QTL) for carcass weight and back fat thickness in Angus cattle (http://www.animalgenome.org/cgi-bin/gbrowse/cattle/#search).
The NOV gene was first identified as an overexpressing gene in virus-induced avian nephroblastoma (Joliot et al., 1992), and orthologs were later isolated from Xenopus, rat, mouse, and human (Joliot et al., 1992; Martinerie et al., 1992; Snaith et al., 1996; Ying and King, 1996; Liu et al., 1999; Oberst et al., 1999). NOV encodes a putative secretory protein of 343 to 357 amino acids that contains four conserved modular domains with sequence similarity to insulin-like growth factor-binding protein (Collet and Candy, 1998). The NOV protein is structurally related to a family of early response proteins that probably play a role in cell growth regulation. It is a member of the growing family of secreted regulatory proteins termed CCN (CYR61/CTGF/NOV) (Bork, 1993). Some CCN proteins have been demonstrated to possess growth factor-like activity and to regulate cell growth and tissue formation. For example, connective tissue growth factor (CTGF) is a matrix-associated heparin binding protein that mediates cell proliferation, migration, and adhesion (Grotendorst, 1997; Moussad and Brigstock, 2000). Compared with CTGF, the biological activity of the NOV gene remains poorly understood. To date, no polymorphisms have been reported in the NOV gene in farm animals.
In the current study, we examined the NOV gene as one of the most promising candidate genes related to meat production in beef cattle. We performed extensive NOV gene screening by resequencing to detect polymorphisms and then examined their genetic associations with carcass traits. Here, we present four polymorphisms identified in the NOV gene as well as an analysis of their associations with carcass yield traits in Korean cattle.
Korean cattle genomic DNA samples were obtained from 429 steers produced from 76 sires used in the progeny testing program at the National Institute of Animal Science (NIAS) in Korea. All steers were fed for 731.39±16.53 d under a tightly controlled feeding program at the Daekwanryeong and Namwon branches of NIAS. They were weaned at a mean age of 3 months and fed with 30% concentrates and 70% roughage until they were 6 months old. After 6 months of age, they were fed concentrates consisting of 15% crude protein (CP) and 71% total digestible nutrients (TDN) until they were 14 months old, followed by 13% CP and 72% TDN until 20 months, and 11% CP and 73% TDN until 24 months of age. Roughage was offered ad libitum, and steers had free access to fresh water during the entire period. Live weights (LW) were determined before slaughter. The mean live weight was 539.95±51.96 kg. Yield grades for carcasses were determined by the cold carcass weight (CW). Dressing percentage (DP) was determined using CW as a proportion of LW. After the carcasses were chilled for 24 h, CW was measured. Then the left side of each carcass was cut between the last rib and the first lumbar vertebrae to determine back fat thickness (BF) (APGS, 1995). The mean carcass trait values were 311.44±33.20 kg for CW, 57.64±1.83% for DP, and 0.70±0.28 cm for BF.
We sequenced the full 8.4-kb NOV gene, including the promoter region (1.5 kb), to discover variants among 24 unrelated Korean cattle, using an ABI PRISM 3730XL DNA analyzer (Applied Biosystems, Foster City, CA, USA). For amplification and sequencing analysis, 16 primer sets were designed based on GenBank sequences (Ref. Genome Seq.; NOV: NC_007312). Primer information is provided in Table 1.
We applied PCR-RFLP methods to detect four SNPs in the NOV gene. Four primer sets flanking DNA fragments of 372 bp, 823 bp, 941 bp, and 933 bp at the mutation sites in Exon 3, Intron 4, and the 3′ UTR region were synthesized. PCR amplification was performed using 20 ng of genomic DNA, the four primer sets (Table 1), and HS Prime Taq polymerase (GENET BIO, Korea). Amplifications were performed in a thermal cycler (PT-200, MJ Research, MA, USA) as follows: 94°C for 10 min, followed by 35 cycles of 94°C for 30 s, 55 to 65°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. The first fragment (372 bp) had a restriction site for HpyCH4; the second fragment (823 bp), for SpeI; the third fragment (941 bp), for HincII; and the fourth fragment (993 bp), for HpyCH4. For restriction enzyme digestion, 5 μl of each PCR product were mixed with 2 units of the appropriate restriction enzyme and incubated at 37°C for 3 h. The digest mixtures were separated in agarose gels containing ethidium bromide for visualization (Figure 3).
A goodness of fit chi-squared test was used to test for Hardy-Weinberg equilibrium (HWE) by comparing the observed number of subjects for each genotype with the expected number of subjects assuming HWE. Genotype distributions at each polymorphic locus were tested for departure from HWE. Associations between individual SNPs and measured carcass traits were determined by regression analysis using the nlme library in the R statistical package (http://www.r-project.org). Trait association analyses were performed using a mixed effect model, treating “sire” as a random effect and “age” at slaughter and genotype as a fixed effect in the model. Other covariates were not available for this analysis. We examined a common measure of linkage disequilibrium (LD) between all pairs of biallelic loci, Lewontin’s D’ (Hedrick, 1987) and r2. Haplotypes and their frequencies were inferred using the algorithm developed by Stephens et al. (2001). A type I error of 5% was used to obtain the Bonferroni corrected P-value. For the haplotype analyses, we fitted the model with the same covariates in a manner similar to that used for the SNP association test.
By resequencing the DNA of 24 unrelated Korean cattle, 19 polymorphisms were identified in the NOV gene: four SNPs in the promoter and exon, and 15 SNPs in the UTR and intron regions. Sequence variants were verified by chromatography (Figure 1). The results showed >99% homology with the sequence reported in GenBank (NC_007312). The locations and allele frequencies of polymorphisms are presented in Table 2 and Figure 2. Pairwise linkage analysis with DNA from the 24 unrelated Korean cattle used for resequencing demonstrated that five sets of polymorphisms were in absolute LD (|D’| = 1 and r2 = 1) in the NOV gene (Figure 2). For the subsequent large-scale association analysis, SNPs were selected based on the following criteria: i) location (exon and promoter polymorphisms were preferred); ii) frequency of minor allele (frequency, 0.05); iii) LD, such that among polymorphisms in absolute LD (r2 = 1), only one was selected; and iv) restriction enzyme site consideration. Among the 19 polymorphisms in the NOV gene, four SNPs (g.1952A>G, g.4590T>G, g.7748T>C, g.8326A>C) were genotyped on a larger scale, in 429 Korean cattle (Table 2).
Large-scale genotyping was performed using the PCR-RFLP method. The PCR-RFLP analysis verified unique binding patterns, showing DNA fragments of different mobilities by agarose gel electrophoresis (Figure 3). Restriction enzyme digestion of the PCR products with HpyCH4IV resulted in fragments of 303 and 69 bp for the G allele, and a 372-bp uncut fragment for the A allele in the g.1952A>G polymorphism. The g.4590T>G SNP was cleaved by SpeI into 621- and 202-bp fragments for the T allele, while a fragment of 823 bp remained uncut for the G allele. For the g.7748T>C polymorphism, digestion with HincII created fragments of 471 and 470 bp for the T allele, and a 941-bp uncut fragment was observed for the C allele. For the g.8326A>C polymorphism, digestion with HpyCH4IV gave 622- and 311-bp fragments for the C allele, while a 933-bp fragment remained for the A allele. No significant departure from HWE was found for any of the alleles (Table 2). The G allele of the g.1952A>G SNP had a low frequency, resulting in low genetic variability with a minor allele frequency of 0.186 and heterozygosity of 0.303. By comparison, the minor allele frequencies for the other SNPs ranged from 0.346 to 0.375, and heterozygosity was 0.453 to 0.469.
Associations of NOV gene polymorphisms with carcass traits were analyzed using the mixed effect model with sire and age as covariates. Sire was treated as a random effect and age as a fixed effect. The obtained p-values were corrected for multiple testing using the Bonferroni correction method (p>Padjusted = 0.0125). The associations between the genotypes and the carcass phenotypes are listed in Table 3.
Statistical analyses revealed that the g.2005A>G polymorphism showed a strong association with BF (p = 0.0001), a weak association with CW (p = 0.036), and no association with LW or DP. The A allele was associated with increased BF (regression coefficient, −0.1) and CW (regression coefficient, −6.3). This result is in agreement with a study by Ferrell and Jenkins (1984), who demonstrated a correlation between CW and BF. The g.4643T>G genotype, which is intronic, was weakly, but significantly, associated with LW (p = 0.029), CW (0.016), DP (0.037), and BF (0.029); however, when Bonferroni corrections were strictly adopted, the respective p-values did not retain significance. It is possible that intronic polymorphisms could influence gene function by altering donor and acceptor splice sites or nearby regions, as well as regulatory motifs within introns.
Two of the polymorphisms located in the 3′ UTR (g.7801T>C and g.8379A>C) showed very strong association with CW. In addition to its significant association with CW (p = 0.012), the g.7801T>C polymorphism had a weak association with LW (p = 0.029), DP (0.037), and BF (0.029). The regression coefficient indicated that the T allele was related to an increase in CW (regression coefficient, 6.0). The g.8379A>C polymorphism was strongly associated with both CW (p = 0.008) and LW (p = 0.012), weakly associated with BF (p = 0.048), and not associated with DP. The A allele of this SNP was related to increased LW and CW (regression coefficients, 9.5 and 6.6, respectively). These results are in agreement with a previous study on Korean cattle that reported a QTL for CW near the RM192 marker, which is close to the NOV gene on BTA14 (Lee et al., 2011). Although the mechanisms by which polymorphisms in the UTR would affect traits are not currently understood, our analysis clearly indicates that, even after correction for multiple testing, two polymorphisms in the UTR influence CW in Korean cattle.
For haplotype-based association analysis, three major haplotypes (frequency, >0.05) were constructed in the NOV gene (Tables 4 and 5). Of nine reconstructed haplotypes, three (HAPLO1, HAPLO2, and HAPLO3) were predominant (total frequency, 94%) in the 429 Korean cattle tested. Other haplotypes were rare and were not analyzed. HAPLO1 was not significantly associated with carcass traits, whereas HAPLO2 showed significant associations with LW, CW, and BF (p = 0.036, 0.021, and 0.041, respectively). HAPLO3 was significantly associated with increased BF (p = 0.00005) and weakly significantly associated with increased LW, CW, and DP (p = 0.055, 0.021, and 0.023, respectively) (Table 3). The estimated two-haplotype effect was larger than the effect of either locus alone; this suggests that none of these SNPs is the causal mutation, but rather that another undetected polymorphism within this haplotype is causing phenotypic variation.
A number of potential candidate genes containing genetic variants have been selected to identify associations with economically important traits based on physiological and biochemical mechanisms. In terms of biochemical mechanisms, the NOV gene belongs to the CCN family, which comprises multifunctional secreted proteins that act as cellular matrix regulators. The extracellular protein products of the CCN gene family, which are approximately 40 kDa in size, regulate numerous biological processes, including differentiation, migration, proliferation, and cell adhesion (Katsube et al., 2009). Although the mechanisms by which alternative genotypes in the UTR and introns could be associated with CW and BF are not currently understood, it is now widely acknowledged that non-coding portion of genomes play crucial roles. Polymorphisms within introns could influence gene function by affecting donor or acceptor splice sites or nearby regions, or regulatory motifs within introns. UTRs are involved in many post-transcriptional regulatory pathways that control mRNA localization, stability, translation efficiency, and initiation of protein synthesis. For UTR and intronic SNPs, although the allele itself may be functional and directly affect the expression of the phenotype, it is more likely that the allele is in linkage disequilibrium with another allele at a nearby locus and that allele is the true causal allele.
The QTL for carcass traits that is located between markers RM192 and BMS947 on bovine chromosome 14 contains many functional candidate genes related to body weight and fat synthesis in beef cattle, as verified with the NOV gene (Lee et al., 2011). A suggested QTL for CW and BF has been reported to occur near 50 cM on BTA14 in Korean cattle (Wibowo et al., 2008). Other QTL studies also have reported that this region on BTA14 is related to BF (Casas et al., 2000; McClure et al., 2010).
In summary, we hypothesized that NOV gene polymorphisms are associated with carcass traits. The results of our study demonstrate that four polymorphisms (two in the 3′ UTR, one exonic, and one intronic) in the NOV gene are associated with LW, CW, and/or BF. Although the mechanisms involved in these associations are not currently understood, this study clearly indicates that NOV gene polymorphisms have an effect on LW, CW, and BF in beef cattle. These findings represent an important step for the successful application of the NOV gene in marker assisted selection programs for commercial beef populations.
Figure 2.
Map of SNPs in the NOV gene on bovine chromosome 14. The first base of the translational site is denoted as nucleotide +1. An asterisk (*) indicates polymorphisms genotyped in Korean cattle (n = 429). The minor allele frequency is based on 24 sequencing samples only, which is different from the minor allele frequency of absolutely linked SNPs genotyped in the larger population.
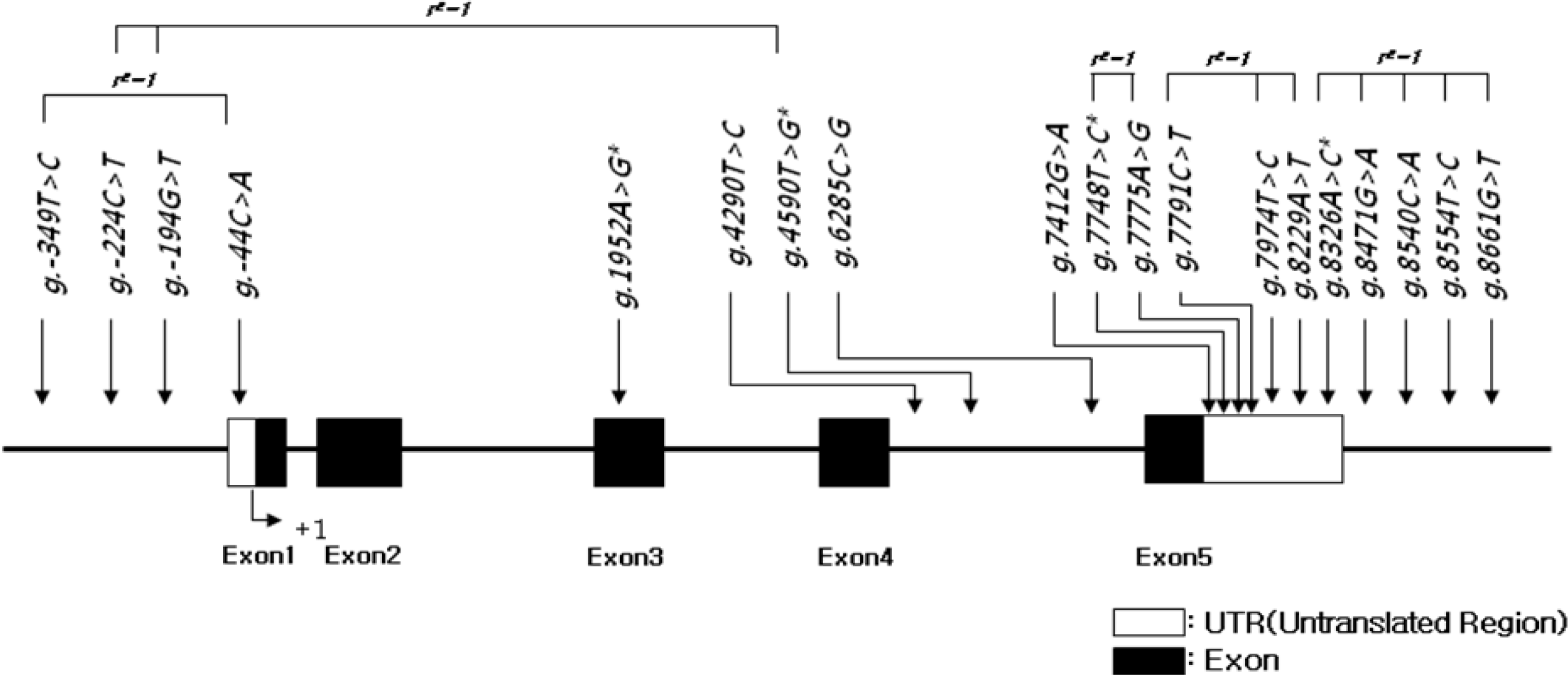
Figure 3.
Sequence chromatograms and PCR-RFLP patterns of four SNPs detected in the bovine NOV gene. The left lane in each gel image is a standard size marker (100 bp DNA ladder).
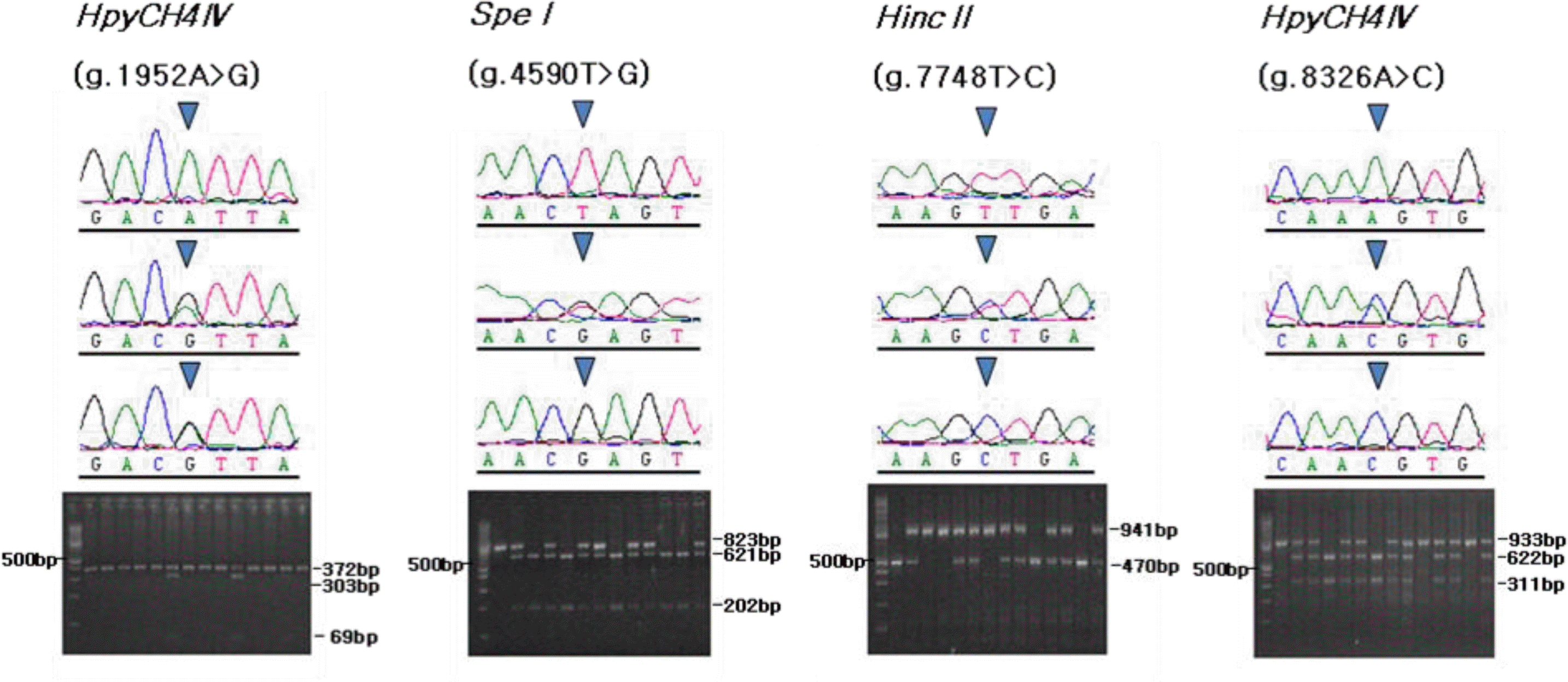
Table 1.
Primer list for SNP discovery and genotyping of NOV gene
Table 2.
Genotype, minor allele frequency (MAF), heterozygosity (He), and Hardy-Weinberg equilibrium (HWE) of 19 polymorphisms in the NOV gene
Table 3.
Analysis of associations between four polymorphisms in the NOV gene and carcass traits in Korean cattle
| Trait | Loci | Position | Genotype | p | R | ||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| C/C | C/R | R/R | |||||
|
|
|||||||
| N(LSMEAN±SE) | N(LSMEAN±SE) | N(LSMEAN±SE) | |||||
| LWT | g.2005A>G | Exon3 | 278(538.0±3.1) | 120(544.2±4.4) | 15(550.7±11.2) | 0.067 | −8.43 |
| g.4643T>G | Intron4 | 178(543.8±3.6) | 184(540.0±4.1) | 57(527.5±6.4) | 0.029 | 8.00 | |
| g.7801T>C | 3′UTR | 158(544.0±3.6) | 191(536.6±4.0) | 59(529.7±6.8) | 0.024 | 8.34 | |
| g.8379A>C | 3′UTR | 177(545.1±3.4) | 200(538.7±4.1) | 49(524.3±6.7) | 0.012* | 9.54 | |
| HAPLO1 | - | 146(539.9±4.3) | 192(536.5±3.5) | 91(547.4±5.1) | 0.434 | −2.65 | |
| HAPLO2 | - | 191(543.0±3.4) | 191(540.3±4.1) | 47(526.4±5.1) | 0.036 | 7.84 | |
| HAPLO3 | - | 299(537.1±3.1) | 116(545.7±4.3) | 14(554.3±11.4) | 0.055 | −8.86 | |
| CW | g.2005A>G | Exon3 | 278(309.8±2.0) | 120(314.6±2.9) | 15(322.9±7.1) | 0.036 | −6.28 |
| g.4643T>G | Intron4 | 178(314.1±2.3) | 184(311.6±2.7) | 57(302.4±4.2) | 0.016 | 5.70 | |
| g.7801T>C | 3′UTR | 158(314.7±2.3) | 191(309.9±2.6) | 59(303.2±4.4) | 0.012* | 6.02 | |
| g.8379A>C | 3′UTR | 177(314.9±2.1) | 200(311.0±2.6) | 49(298.9±4.2) | 0.008* | 6.57 | |
| HAPLO1 | - | 146(311.6±2.8) | 192(309.1±2.4) | 91(316.1±3.2) | 0.452 | −1.64 | |
| HAPLO2 | - | 191(313.5±2.2) | 191(312.1±2.7) | 47(300.1±4.3) | 0.021 | 5.56 | |
| HAPLO3 | - | 299(309.3±2.0) | 116(315.3±2.9) | 14(326.0±6.8) | 0.021 | −6.87 | |
| DP | g.2005A>G | Exon3 | 278(57.5±0.1) | 120(57.8±0.2) | 15(58.6±0.5) | 0.053 | −0.30 |
| g.4643T>G | Intron4 | 178(57.8±0.1) | 184(57.6±0.1) | 57(57.3±0.2) | 0.037 | 0.26 | |
| g.7801T>C | 3′UTR | 158(57.8±0.1) | 191(57.7±0.1) | 59(57.2±0.2) | 0.042 | 0.26 | |
| HAPLO1 | - | 146(57.7±0.2) | 192(57.6±0.1) | 91(57.7±0.2) | 0.697 | −0.04 | |
| HAPLO2 | - | 191(57.7±0.1) | 191(57.7±0.1) | 47(57.0±0.3) | 0.089 | 0.21 | |
| HAPLO3 | - | 299(57.5±0.1) | 116(57.7±0.2) | 14(58.8±0.5) | 0.023 | −0.35 | |
| BF | g.2005A>G | Exon3 | 278(6.8±0.2) | 120(7.5±0.3) | 15(8.6±0.9) | 0.0001* | −0.96 |
| g.4643T>G | Intron4 | 178(7.3±0.2) | 184(6.9±0.2) | 57(6.6±0.3) | 0.029 | 0.44 | |
| g.7801T>C | 3′UTR | 158(7.4±0.2) | 191(6.9±0.2) | 59(6.6±0.3) | 0.020 | 0.47 | |
| g.8379A>C | 3′UTR | 177(7.2±0.2) | 200(7.0±0.2) | 49(6.5±0.3) | 0.048 | 0.41 | |
| HAPLO1 | - | 146(7.2±0.2) | 192(7.0±0.2) | 91(7.0±0.3) | 0.495 | 0.12 | |
| HAPLO2 | - | 191(7.2±0.2) | 191(7.0±0.2) | 47(6.5±0.4) | 0.041 | 0.41 | |
| HAPLO3 | - | 299(6.8±0.2) | 116(7.5±0.3) | 14(8.9±0.9) | 0.00005* | −1.00 | |
REFERENCES
APGS. 1995. Report of business for animal products grading.: Animal products garding system. National Livestock Cooperatives Federation; Korea:
Bork P. 1993. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett 327:125–130.


Casas E, Shackelford SD, Keele JW, Stone RT, Kappes SM, Koohmaraie . 2000. Quantitative trait loci affecting growth and carcass composition of cattle segregating alternate forms of myostatin. J Anim Sci 78:560–569.


Collet C, Candy J. 1998. How many insulin-like growth factor binding proteins? Mol Cell Endocrinol 139:1–6.


Ferrell CL, Jenkins TG. 1984. Relationships among various body components of mature cows. J Anim Sci 58:222–233.


Grotendorst GR. 1997. Connective tissue growth factor: a mediator of TGF-beta action on fibroblasts. Cytokine Growth Factor Rev 8:171–179.


Hermesmeyer GN, Berger LL, Nash TG, Brandt RT. 2000. Effects of energy intake, implantation, and subcutaneous fat end point on feedlot steer performance and carcass composition. JAnim Sci 78:825–831.

Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, Perbal B. 1992. Proviral rearrangements and overexpression of a new cellular gene (nov) in myeloblastosis-associated virus type 1-induced nephroblastomas. Mol Cell Biol 12:10–21.



Katsube K, Sakamoto K, Tamamura Y, Yamaguchi A. 2009. Role of CCN, a vertebrate specific gene family, in development. Dev Growth Differ 51:55–67.


Lee SH, van der Werf JH, Kim NK, Lee SH, Gondro C, Park EW, Oh SJ, Gibson JP, Thompson JM. 2011. QTL and gene expression analyses identify genes affecting carcass weight and marbling on BTA14 in Hanwoo (Korean Cattle). Mamm. Genome 22:589–601.

Liu C, Liu XJ, Crowe PD, Kelner GS, Fan J, Barry G, Manu F, Ling N, De Souza EB, Maki RA. 1999. Nephroblastoma overexpressed gene (NOV) codes for a growth factor that induces protein tyrosine phosphorylation. Gene 238:471–478.


Martinerie C, Viegas-Pequignot E, Guenard I, Dutrillaux B, Nguyen VC, Bernheim A, Perbal B. 1992. Physical mapping of human loci homologous to the chicken nov proto-oncogene. Oncogene 7:2529–2534.

McClure MC, Morsci NS, Schnabel RD, Kim JW, Yao P, Rolf MM, McKay SD, Gregg SJ, Chapple RH, Northcutt SL, Taylor JF. 2010. A genome scan for quantitative trait loci influencing carcass, post-natal growth and reproductive traits in commercial Angus cattle. Anim Genet 41:597–607.


Moon SS, Hwang IH, Jin SK, Lee JG, Joo ST, Park GB. 2003. Carcass traits determining quality and yield grades of Hanwoo steers. Asian Australas J Anim 16:1049–1054.

Moussad EE, Brigstock DR. 2000. Connective tissue growth factor: what’s in a name? Mol Genet Metab 71:276–292.


Oberst C, Hartl M, Weiskirchen R, Bister K. 1999. Conditional cell transformation by doxycycline-controlled expression of the MC29 v-myc allele. Virology 253:193–207.


Snaith MR, Natarajan D, Taylor LB, Choi CP, Martinerie C, Perbal B, Schofield PN, Boulter CA. 1996. Genomic structure and chromosomal mapping of the mouse nov gene. Genomics 38:425–428.


Stephens M, Smith NJ, Donnelly P. 2001. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989.



- TOOLS
-
METRICS

-
- 1 Crossref
- 0 Scopus
- 7,604 View
- 41 Download
- Related articles





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




