 |
 |
Abstract
The use of Korean native chicken is increasing, and the discovery of new genetic resources is very important from both economic and genetic conservation points of view. In this study, mtDNA D-loop sequences from 272 privately-owned Korean native chickens from a Hyunin farm were investigated. Seventeen nucleotide substitutions were identified from the sequence analysis and they were classified as 6 haplotypes. Previously investigated haplotypes in five Korean native chicken populations have been compared with the Hyunin chicken population. The results indicated that two haplotypes, H10 and H15, in the Hyunin chicken population were not previously identified in other Korean native chicken populations, representing 33.09% (90/272) and 1.1% (3/272) of the Hyunin population, respectively. On the other hand, four other haplotypes were identical to those of a previous study of Korean native chicken populations. This result is indicative of conservation strategies of Hyunin chicken populations for expanding the genetic diversity in the Korean native chicken population.
Increased research and genetic profiling of native species is an important component of modern perspectives of genetic diversity conservation. There is also an increasing demand for native chicken meat currently, even though 90% of chicken consumed in Korea is from imported breeds (MIFAFF, 2009). Five lines of Korean native chicken populations were developed in the 1990’s, and this breeding was mainly controlled by a national institution called National Institute of Animal Science (NIAS) in Korea. However, there are still native chicken breeds available in the private farms, and one of them, the Hyunin population, is used in this study for comparison with the primary Korean native chicken populations for which we have previous genetic information.
Maternally-inherited mitochondrial DNA (mtDNA) sequences have been successfully used for genetic diversity studies of avian and domestic animals (Komiyama et al., 2003, 2004; Liu et al., 2006; Odahara et al., 2006; Sasazaki et al., 2006; Lei et al., 2007; Wang et al., 2007; Li et al., 2008). In the mitochondrial genome, coding genes are predominantly used for biodiversity studies of many different species, whereas, the non-coding control region is used for the inter-specific population studies (Baker and Marshall, 1997; Moore and Defilippis, 1997). MtDNA has the advantages of the absence of recombination and rapid nucleotide substitutions compared with genomic DNA sequences (Aquadro and Greenberg, 1983; Lansman et al., 1983; Cann et al., 1984). Chicken D-loop sequences were previously determined to investigate the origin of breeds or species, and various haplotypes were reported (Komiyama et al., 2003, 2004; Liu et al., 2006; Oka et al., 2007). Previously, three SNPs of D-loop region were found to have significant discrimination power for two commercial native chicken populations in Korea (Hoque et al., 2011). Also, the D-loop hypervariable region of mtDNA was used to determine the relationships between Korean native chickens and other chicken breeds (Hoque et al., 2009).
In this study, the chicken mtDNA D-loop region was used for the demarcation of haplotypes in the privately-owned Hyunin native chicken population, and was compared with those of other Korean native chicken breeds, which were previously developed in NIAS. This Hyunin population has distinct feather colors and they were classified as eleven colors. Some of these (red, yellow, black, gray and white) are the same colors identified in Korean native chickens at NIAS. These results provide a basis for an appropriate conservation breeding program for the Korean native chicken.
A total of 272 native chicken samples were collected from a single flock in a Hyunin chicken farm. Genomic DNAs were extracted from blood samples using a manual extraction method (Miller et al., 1998) applied to a large volume, which was stored at −20°C until use. For the comparison of haplotypes, previous D-loop sequence data from two commercial chicken populations Hanhyup (40 samples) and Yellim (37 samples) and three Korean native chicken breeds (88 samples) were included in the analysis (Hoque et al., 2011).
PCR was used to amplify a 591 bp fragment in the D-loop hypervariable region in mtDNA by the following primer pair (Forward: 5′- AGGACTACGGCTTGAAAAGC -3′ and Reverse: 5′-ATGTGCCTGACCGAGGAACCAG -3′). The PCR reactions included approximately 100 ng of genomic DNA, 2.5 μl of 10× buffer (Tris-HCl (pH 9.0), PCR enhancers, (NH4)2SO4, 20 mM MgCl2), 2.0 μl of 10 mM dNTPs mixture (2.5 mM each of dATP, dCTP, dGTP and dTTP), 1 μl of 10 pM of each primer and 1 U HS Prime Taq (GeNet Bio, Korea) in a 25 μl reaction volume. PCR was performed in a My-Genie96 Thermal Block (Bioneer) with an initial denaturation step at 94°C for 10 min, followed by 35 cycles of 30 s at 94°C, 30 s at 61°C, 40 s at 72°C and a final extension step at 72°C for 10 min. The PCR products for mtDNA were electrophoresed on 1.5% agarose gels with ethidium bromide, and DNA bands were visualized under ultraviolet light. Purification of PCR products was performed using an Accuprep®PCR purification kit (Bioneer) according to the manufacturer’s instructions. Purified PCR products were also confirmed using agarose gels for sequencing. All the purified PCR products were sequenced by Genotech (www.genotech.co.kr) commercial company.
The chicken mtDNA D-loop nucleotide sequence data were aligned using the ClustalW program (Thompson et al., 1994). Nucleotide replacement export data from mtDNA were carried out in haplotype sequences by using MEGA5 (Tamura et al., 2011). The number of haplotypes, nucleotide variable site, haplotype diversity and nucleotide diversity (Nei, 1982) were calculated using DnaSP V.5.10 (Rozaset al., 2003). A rectangular Kimura 2-parameter model neighbor-joining (NJ) phylogenetic tree with 1000 bootstrap replications was constructed using MEGA5. Also, an outline for median-joining network profiles with positional SNPs for the differentiation of haplotypes by using NETWORK4610 software (Bandelt et al., 1999).
Mitochondrial D-loop sequence variations were used to calculate the number of variable sites (S), number of haplotypes (h), haplotype diversity (Hd) and nucleotide diversity (Pi) among the chicken populations (Table 1). Analysis of 272 mtDNA D-loop sequence data from the Hyunin population revealed six haplotypes, which were comprised of seventeen nucleotide substitutions. All of the six mtDNA haplotypes for the Hyunin chicken population that were discovered are shown in Table 2. Based on the statistical analysis, the identified Hd and Pi were 0.768 and 0.0083, respectively, in Hyunin chickens. Based on the previous analysis of 40 sequences from Hanhyup commercial chicken population, 15 variable sites were identified which have 5 haplotypes with 0.778 of Hd and 0.0068 of Pi. Interestingly, 37 mtDNA D-loop sequences were conserved in the Yellim commercial chicken population (Hoque et al., 2011). Moreover, three Korean native chicken breeds have been compared with the three Korean native chicken populations, dominated by black, red, and yellow, respectively. The black Korean native chicken (KB) breed was observed to be highly variable in terms of S, h, Hd and Pi for 22, 9, 0.823 and 0.0113, respectively. Red Korean native chicken (KR) breeds also had a high nucleotide substitution of 20, and 6 haplotypes which used to calculate a Hd value of 0.662 and a Pi value of 0.0093. On the other hand, the yellow Korean native chicken (KY) breed had a comparatively low number of variable sites of 10 and 4 haplotypes, from which a Hd and Pi of 0.591 and 0.0077 were calculated, respectively.
A total of 28 nucleotide substitutions were observed among the chicken populations which represent 15 haplotypes (Table 3). Among these, 6 haplotypes were identified in the Hyunin population. Of these, two haplotypes were unique to the Hyunin population. On the other hand, four haplotypes were mixed with other Korean chicken populations. Of these, four haplotypes (Hnhap5, Hnhap4, Hnhap2 and Hnhap1) were highly related with the KR breed (25 samples) which represents H2, H9, H11 and H12, respectively. In the case of the KB breed, only six chickens shared three Hyunin haplotypes (Hnhap5, Hnhap4 and Hnhap2) which are the H2, H9 and H11, respectively. Only one haplotype (Hnhap5) was shared with the KY breed (3 samples). Based on the haplotype sharing among the Korean native chicken breeds, the Hyunin chicken population is very closely related to the KY chicken breed. Two chicken populations (Hanhyup and Yellim) had Hnhap4 representing the H9 haplotype. On the other hand, specific haplotypes for Hyunin population have contained 33.09% of Hnhap3 (90 samples) and 1.1% of Hnhap6 (3 samples) which represent H10 and H15 haplotypes, respectively. The H10 haplotype, containing the D-loop nucleotide substitution C210T, is unique to the Hyunin chicken breed. Also, three SNP positions, G212A, C246T and C315T, in the D-loop region are specific for H15 discrimination of the Hyunin breed. Previously, we successfully differentiated two commercial chicken populations by the selected three D-loop SNPs at positions C225T, A239G and C243T (Hoque et al., 2011). Also, it was demonstrated that allele-specific SNP typing in mtDNA D-loop region at the five SNP sites (C310T, A342G, C446T, A686G and C1213T) were SNP haplotypes that can be used for identification of the origins of chicken meat (Harumi et al., 2011). The D-loop region in mtDNA may also play a vital role in the measurement of diversity, and in evaluating the population structure of native Korean chickens.
The estimation of network profiles was traced using 15 haplotypes in the D-loop nucleotide positions of native chicken populations (Figure 1). In our studies, haplotype H10 was found to specifically represent the Hyunin population that differentiated H9 by the C210T SNP. Also, the H15 haplotype for the Hyunin population is distinguished from other haplotypes by the three nucleotide positions at 212, 246, and 315. Moreover, the H9 Hyunin Haplotype is differentiated with H12 by the four nucleotide positions at 291, 313, 367 and 370. Besides, the H12 haplotype is differed from the H11 haplotype by 199 nucleotide position for the Hyunin haplotype. Only haplotype H2 is markedly different from other Hyunin haplotypes. These specific SNPs observed in the D-loop region will provide valuable markers for the discrimination of Korean native chicken breeds.
The phylogenetic analysis among the chicken populations was conducted for identifying relationships between Hyunin chicken populations and previously identified Korean native chicken breeds (Figure 2). In our phylogenetic analysis, haplotypes were clearly differentiated among the two groups. Five haplotypes for the Hyunin population were clustered together in one clade (lower clade in Figure 2) and only one haplotype was contained in another clade (upper clade in Figure 2), while, three haplotypes in the lower clade were mostly related with the KR breed. The internal Hyunin haplotype Hnhap4 is also related with two commercial chicken populations and the KB haplotype. In addition, Hnhap2 contains the KB haplotype. Interestingly, Hnhap3 and Hnhap6 are completely separated from other haplotypes, which was determined through network profile analysis. On the other hand, Hnhap5 in the upper clade is closely related with KR and KB haplotypes. Our phylogenetic analyses also gives some idea as to the relationship among Korean native chicken populations.
In this study, we investigated the D-loop SNPs and haplotypes for the identification of Korean native chicken populations in a Hyunin private farm. Two previously unidentified haplotypes were investigated, and the phylogenetic relationships were observed by comparing them with haplotypes of other chicken breeds. The results indicate that the private farms still hold valuable genetic resources which had not been previously identified. To maintain this valuable native chicken population, an appropriate conservation breeding program is ultimately required, and should be conducted with the aid of molecular genetic techniques.
ACKNOWLEDGMENTS
This study was carried out with the support of “FTA Agriculture Research Project (Project No. PJ9070112011)”, RDA, Republic of Korea.
Figure 1
Network profiles of the 15 haplotypes from the six Korean native chicken populations. The links are labeled by the nucleotide positions. The order of the mutations on the branch is arbitrary. The median vectors (mv) are nucleotide junctions.
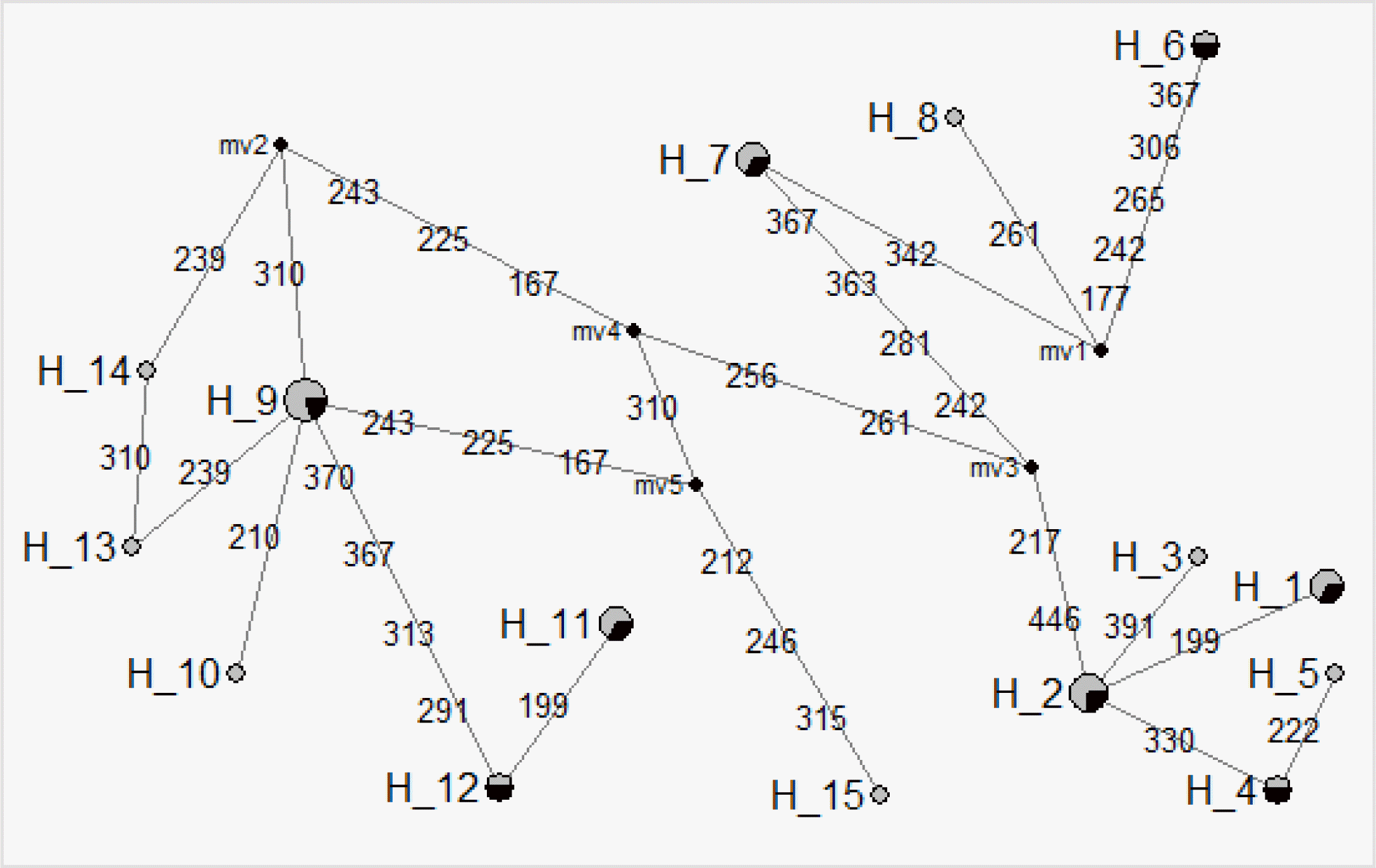
Figure 2
The neighbor-joining phylogenetic tree using Kimura 2-parameter model with 1000 bootstrap replications in six Korean native chicken populations.
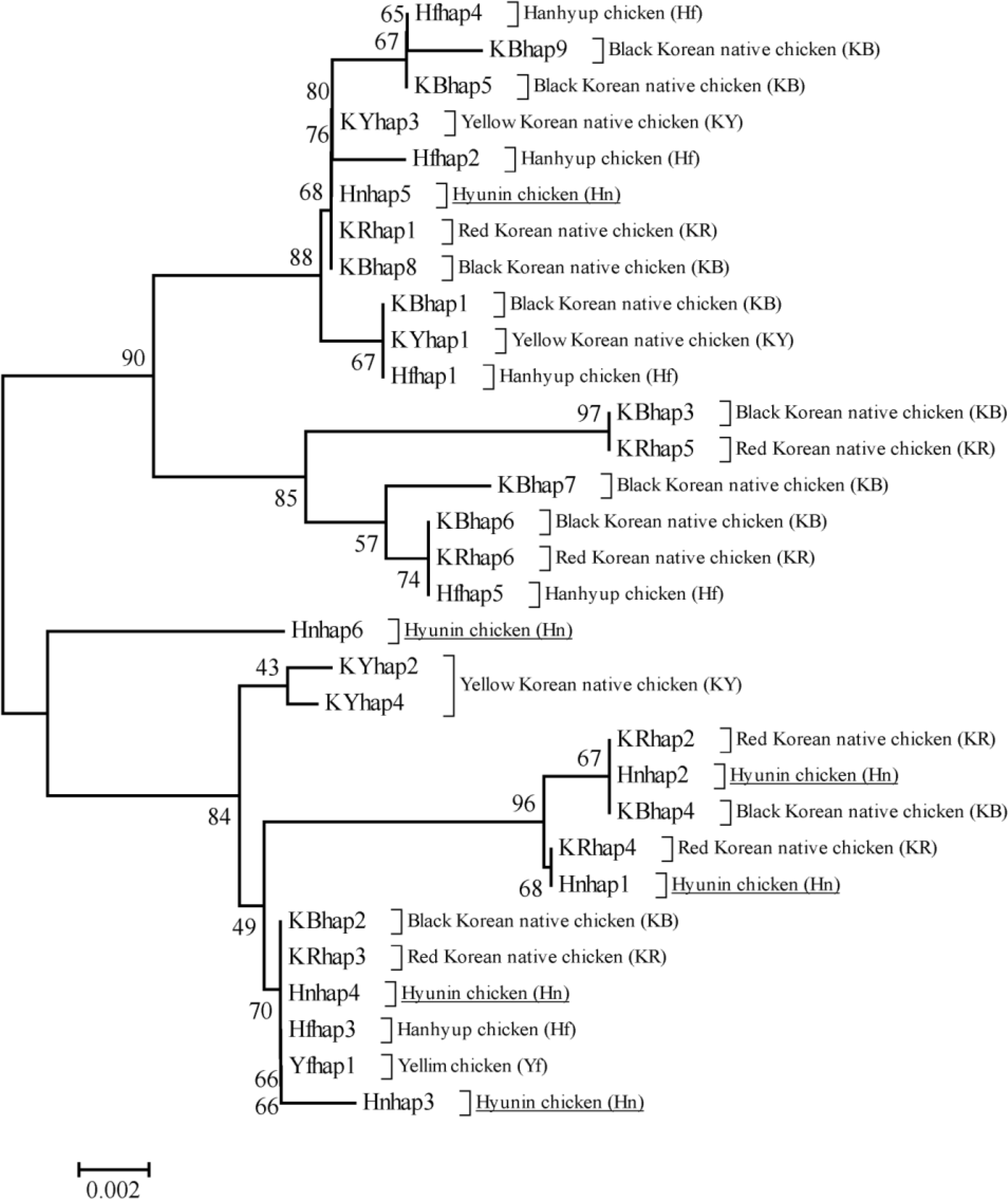
Table 1
The calculated number of variable sites (S), number of haplotype (h), haplotype diversity (Hd) and nucleotide diversity (Pi) using mitochondrial D-loop sequence polymorphisms for the Korean native chicken populations
S = Number of variable sites, h = Number of haplotype, Hd = Haplotype diversity, Pi = Nucleotide diversity.
Except for the sequences from Hyunin, the sequence data were obtained from Hoque et al. (2011).
Table 2
The identified haplotypes with the mitochondrial D-loop sequence polymorphisms in Hyunin chicken
Table 3
The identified haplotypes with the mitochondrial D-loop sequence polymorphisms among the Korean native chicken populations
REFERENCES
Aquadro CF, Greenberg BD. 1983. Human mitochondrial DNA variation and evolution: Analysis of nucleotide sequences from seven individuals. Genetics 103:287–312.




Baker AJ, Marshall HD. 1997. Mitochondrial control region sequences as tools for understanding evolution. Avian molecular evolution and systematic. Mindell DP, editorAcademic press; San Diego: p. 51–82.

Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48.


Cann RL, Brown WM, Wilson AC. 1984. Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics 106:479–499.




Harumi T, Sano A, Minematsu T, Naito M. 2011. Allele-specific PCR typing and sequencing of the mitochondrial D-loop region in four layer breeds. Anim Sci J 82:223–226.


Hoque MR, Jung KC, Park BK, Choi KD, Lee JH. 2009. Genetic variability of mtDNA D-loop region in Korean native chickens. Korean J Poult Sci 37:323–328.

Hoque MR, Lee SH, Jung KC, Kang BS, Park MN, Lim HK, Choi KD, Lee JH. 2011. Discrimination of Korean native chicken populations using SNPs from mtDNA and MHC polymorphisms. Asian-Aust. J Anim Sci 24:1637–1643.


Komiyama T, Ikeo K, Gojobori T. 2003. Where is the origin of the Japanese gamecock? Gene 317:195–202.


Komiyama T, Ikeo K, Gojobori T. 2004. The evolutionary origin of long-crowing chicken: its evolutionary relationship with fighting cocks disclosed by the mtDNA sequence analysis. Gene 333:91–99.


Lansman RA, Avise JC, Huettel MD. 1983. Critical experimental test of the possibility of ‘paternal leakage’ of mitochondrial DNA. Proc Natl Acad Sci USA 80:1969–1971.



Lei CZ, Zhang W, Chen H, Lu F, Ge QL, Liu RY, Dang RH, Yao YY, Yao LB, Lu ZF, Zhao ZL. 2007. Two maternal lineages revealed by mitochondrial DNA dloop sequences in Chinese native water buffaloes (Bubalus bubalis). Asian-Aust. J Anim Sci 20:471–476.

Li JL, Shi Y, Fan C, Manglai D. 2008. mtDNA diversity and origin of Chinese Mongolian horses. Asian-Aust. J Anim Sci 21:1696–1702.

Liu YP, Wu GS, Yao YG, Miao YW, Luikart G, Baig M, Pereira AB, Ding ZL, Palanichamy MG, Zhang YP. 2006. Multiple maternal origins of chickens: Out of the Asian jungles. Mol Phylogenet Evol 38:12–19.


MIFAFF. 2009. Primary statistics of food. Agriculture, Forestry and Fisheries; Korea:
Miller SA, Dykes DD, Polesky HF. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215



Moore WS, Defilippis VR. 1997. The window of taxonomic resolution for phylogenies based on mitochondrial cytochrome b. Avian molecular evolution and systematic. Mindell DP, editorAcademic press; San Diego: p. 84–119.

Nei M. 1982. Evolution of human races at the gene level. Prog Clin Biol Res 167–181.
Odahara S, Chung HJ, Choi SH, Yu SL, Sasazaki S, Mannan H, Park CS, Lee JH. 2006. Mitochondrial DNA diversity of Korean native goats. Asian-Aust. J Anim Sci 19:482–485.

Oka T, Ino Y, Nomura K, Kawashima S, Kuwayama T, Hanada H, Amano T, Takada M, Takahata N, Hayashi Y, Akishinonomiya F. 2007. Analysis of mtDNA sequences shows Japanese native chickens have multiple origins. Anim Genet 38:287–293.


Rozas J, Sanchez-DelBarrio JC, Messeguer X. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497.


Sasazaki S, Odahara S, Hiura C, Mukai F, Mannen H. 2006. Mitochondrial DNA variation and genetic relationships in Japanese and Korean cattle. Asian-Aust. J Anim Sci 19:1394–1398.

Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739.



- TOOLS
-
METRICS

- Related articles
-
Genetic Analysis of Haimen Chicken Populations Using Decamer Random Markers2006 November;19(11)





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




