 |
 |
Abstract
Based on a quantitative traits locus (QTL) study using a F2 intercross between Landrace and Korean native pigs, a significant QTL affecting teat numbers in SSC7 was identified. The strong positional candidate gene, TBC1D21, was selected due to its biological function for epithelial mesenchymal cell development. Sequence analysis revealed six single nucleotide polymorphisms (SNPs) in the TBC1D21 gene. Among these, two SNP markers, one silent mutation (SNP01) for g.13,050A>G and one missense mutation (SNP04) for c.829A>T (S277C), were genotyped and they showed significant associations with teat number traits (p value = 6.38E-05 for SNP01 and p value = 1.06E-07 for SNP04 with total teat numbers). Further functional validation of these SNPs could give valuable information for understanding the teat number variation in pigs.
Teat numbers in pigs are an important trait since they determine the mothering ability of the sows. The number of available teats can also affect live-born per litter and weaning weight of piglets, and these traits are related with the mother conditions for the next parity. Usually, most of the reproductive traits have very low heritabilities. However, teat numbers have relatively medium heritability (h2 = 0.32), compared with other reproductive traits, indicating that the individual selection can be applicable to the genetic improvement for this trait (Pumfrey et al., 1980). This also suggests that the identification of the causative mutation(s) in teat numbers will be more feasible than other reproductive traits such as littler size.
Previously, genome scans were carried out for identifying the causative chromosomal locations for the teat numbers in the porcine genome and several quantitative traits locus (QTL) locations were identified. Wada et al. (2000) identified significant QTLs in SSC1 and SSC7 from a Meishan×Göttingen cross population. The QTL information was summarized in the database called “Animal QTLdb” and the database identified 78 QTL regions in 13 autosomes (SSC1, 2, 3, 4, 5, 6, 7, 8, 10, 11, 12, 15, 17) and the X chromosome. Especially, 10 and 9 teat number related QTLs were identified on SSC1 and SSC7, respectively (http://www.animalgenome.org/cgi-bin/QTLdb/SS/index).
Recently, a positional candidate gene on SSC5, PTHLH (parathyroid hormone-like hormone), was investigated for the relationship with teat numbers. However, the results indicated that there was no significant relationship with PTHLH genotype and the teat number phenotype (Martinez-Giner et al., 2011).
In our QTL study using an F2 intercross between Landrace and Korean native pigs, two significant QTLs were identified in SSC1 and SSC7 (data not shown). When we compared these two QTLs, SSC7 QTL was more significant than the QTL on SSC1 and further study was carried out only for the SSC7 QTL. Within the significant QTL region, TBC1D21 was selected as the positional candidate gene based on the possible function of this gene.
Therefore, the aim of this study was to investigate the relationships between SNPs in the positional candidate gene, TBC1D21, and teat number traits in an F2 intercross population between Landrace and Korean native pigs.
Recently, there are relatively increase uses of the Korean native pigs not only for meat purpose but also for the animal model (Oh et al., 2011). The animals used in this study were from a three-generation resource population obtained by crossing Landrace and Korean native pigs (Cho et al., 2011). Briefly, nineteen purebred KNP (eight males and eleven females) were crossed with seventeen purebred Landrace (eight males and nine females). From these crosses, ninety-one F1 progeny and 1,106 F2 progeny (568 males and 538 females) from 79 full-sib families were produced. Teat numbers (total teat numbers, right teat numbers and left teat numbers) were counted in 1,105 F2 progenies (568 males and 537 females) from 79 full-sib families in this resource population Of these, only F2 animals successfully genotyped were included further association analysis in this study.
Initially, 18 F0 animals in the resource population were used to detect the single nucleotide polymorphisms (SNPs). After purifying the PCR products using a PCR purification kit (Bioneer, Korea), the products were sequenced by Cosmo Genetech (http://www.cosmogenetech.com). The DNA sequences were analyzed with the BioEdit program ver. 7.0.0 (Ibis Therapeutics, USA) and the SNPs were identified by a manual search of the electrophoregram results (data not shown).
For the pyrosequencing analysis of two SNPs in the TBC1D21 gene, primer sets and minisequencing primers were used for PCR reactions (Table 1). Reactions were performed using 50 ng of geneomic DNA, 0.4 μM of each primer, 1.5 mM MgCl2, reaction buffer (Genetbio, Korea), 0.2 mM dNTP, and Taq DNA polymerase (Genetbio, Korea) in a 25 μL reaction volume. The PCR condition was 35 cycles of 30 s at 94°C (denaturation), 45 s at 64°C (annealing), 45 s at 72°C (elongation), and 2 min at 94°C (denaturation). The genotype of the PCR products was analyzed using a pyrosequencer (PyroMark, USA).
Student’s-t test was performed to examine the difference between the left and right teat numbers using SAS package ver 9.2 (SAS, USA). Association between teat number traits and the SNP markers were analyzed by a mixed model using the PROC MIXED option implemented in SAS package ver 9.2. The model included sire and dam (nested within sire) as random effects, sex and genotype as fixed effects. Significant differences among least square means of the different genotypes were computed using a contrast test. Unless otherwise stated, significance was set at p<0.05. In addition, Haploview (ver 4.1) was used to analyze Linkage disequilibrium (LD) (Barrett et al., 2005). LD between SNP markers was assessed by calculating a pairwise measure of LD described by Lewontin (Lewontin, 1964).
The mean total teat number in F2 individuals was 13.67 ±0.039 with minimum and maximum values of 10 and 18, respectively. There was no teat number difference between right and left (p = 0.19, Table 2). When we compared the mean teat numbers with another study, similar results were observed in the Landrace breed, which had mean teat numbers of 13.78±1.3. On the other hand, purebred Korean native pigs had a mean of 12.10±1.2, which indicates at least one teat smaller than those of Landrace and current F2 individuals (Seo et al., 1996).
Previous studies reported that the similar QTL positions on SSC7 having significant QTL peak locations between 49.4 and 100.9 cM (Wada et al., 2000; Cassady et al., 2001; Bidanel et al., 2008). However, none of the previous studies investigated the candidate gene(s) in this QTL region. Using the current F2 individuals in this study, we also identified the teat number QTL located in the similar chromosomal region of SSC7 compared with the previous studies (data not shown). In this QTL region, TBC1D21 was selected as the positional candidate gene based on the possible function of this gene.
TBC1D21 gene (TBC1 domain family member 21) is located on human chromosome 15, acts as a GTPase-activating protein for Rab family proteins. Rab family proteins comprise G proteins of the Ras superfamily that belong to a protein superfamily of small GTPases. Approximately 70 types of Rabs have been identified in humans. They are regulators of membrane traffic, including vesicle formation, vesicle movement along actin and tubulin networks, and membrane fusion (Fuchs et al., 2007). Therefore, the TBC1D21 gene, which contains a Rab-GAP (GTPase activation protein) domain, may have a significant effect on the signaling in epithelial mesenchymal interaction during the formation of the mammary gland. Therefore, the TBC1D21 gene was investigated as an important positional candidate gene in this region.
To identify the SNPs of TBC1D21 gene, sequencing was performed. By manual examination of the sequencing result of this gene, we identified six SNPs in exon regions. Among the six sequence variants, two SNPs (SNP01 and SNP04) were selected for genotyping. SNP01 (g.13,050A>G) was revealed as a silent mutation and SNP04 (c.829A>T(S277C)) as a missense mutation. The haploview results indicated that these two SNPs are highly linked, so it was anticipated that the significant results would be very similar each other (Figure 1).
For the efficient genotyping of the identified two SNPs, pyrosequencing was carried out (Figure 2). In case of SNP01, the identified allele frequencies were 0.8 for the A allele and 0.2 for the G allele, and the genotype frequencies were 0.7 (AA), 0.21 (AG), and 0.09 (GG) (Table 3). Also, for SNP04, the allele frequencies identified were 0.84 and 0.16 for the A and T allele, respectively, and the genotype frequency was 0.05, 0.22, and 0.73 for the TT, TA, and AA genotype, respectively (Table 3).
A mixed effect model was used to investigate the effects of SNPs in TBC1D21 gene on teat number in a F2 intercross between KNP and Landrace pigs. The two SNPs that were genotyped were highly significant, indicating a greater significance of SNP04 than SNP01 (Table 4). The identified P value for the total teat numbers was 1.06E-07 and 6.38E-05 for SNP04 and SNP01, respectively. The results indicated that the greater preference of the less frequent homozygous alleles in both SNPs.
In a previous study, the PTHLH gene was selected as the positional candidate gene on SSC5 in the QTL region because this gene contributes to the formation of the nipple (Martinez-Giner et al., 2011). Presently, TBC1D21 gene was selected as the first candidate gene on SSC7 and the two identified genotyped SNPs were significant for teat number traits. Further functional validation of the identified mutations in this gene can greatly contribute for increased teat numbers in sows, which also can be used for improving the productivities in pigs.
ACKNOWLEDGMENTS
This research was supported by a grant from the Technology Development Program (Project no. 110061-03-1-CG000) for Agriculture and Forestry, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea.
Figure 1.
Exonic nucleotide variants and LD block of the TBC1D21 gene. For haplotype analysis, linkage disequilibrium (LD) block between SNP markers was estimated using Haploview v4.2 software.

Figure 2.
Pyrosequencing results and genotypes of the TBC1D21 gene. (A) Pyrosequencing of SNP01, the exon nucleotide g.13,050A>G (B) Pyrosequencing of SNP04, the exon nucleotide g.11,496A>T (c.829A>T(S277C)).
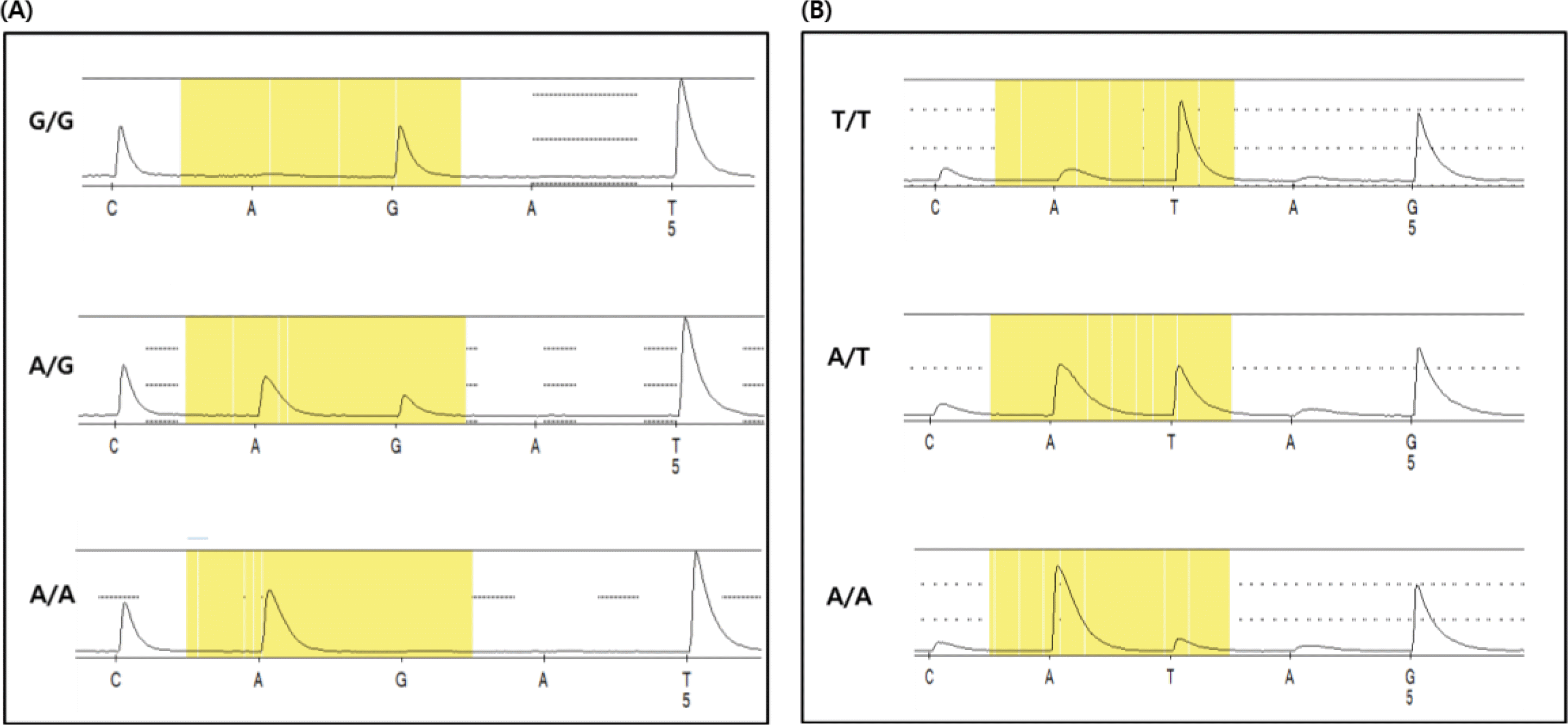
Table 1.
Pyrosequencing primer set for genotyping the SNPs in the TBC1D21 gene
Table 2.
The basic statistics for the teat number traits in F2 intercross between KNP and Landrace pigs
| Phenotype | N | Mean | Standard deviation | Minimum | Maximum |
|---|---|---|---|---|---|
| Total teat number | 1105 | 13.665 | 1.288 | 10 | 18 |
| Left teat number | 1105 | 6.812 | 0.734 | 5 | 9 |
| Right teat number | 1105 | 6.853 | 0.741 | 5 | 9 |
Table 3.
The alllele and genotype frequencies for the SNPs in the TBC1D21 gene
| SNP | Allele frequency | Genotype frequency | |||
|---|---|---|---|---|---|
|
|
|
||||
| SNP01 | 0.2(G) | 0.8(A) | 0.09(GG) | 0.21(GA) | 0.7(AA) |
| g.13,050A>G | |||||
| SNP04 | 0.16(T) | 0.84(A) | 0.05(TT) | 0.22(TA) | 0.73(AA) |
| g.11,496A>T | |||||
| (c.829A>T(S277C)) | |||||
Table 4.
Effect of SNPs in the TBC1D21 gene on teat number traits in F2 intercross between KNP and Landrace pigs
| SNP | Trait |
Genotype
|
p value | ||
|---|---|---|---|---|---|
| GG (SE) | AG (SE) | AA(SE) | |||
| SNP01 | Total teat number | 12.94(0.17)a | 13.44(0.10)b | 13.73(0.07)c | 6.38E-05 |
| g.13,050A>G | Left teat number | 6.58(0.10) | 6.75(0.06) | 6.83(0.04) | 0.08 |
| Right teat number | 6.36(0.10)a | 6.69(0.06)b | 6.90(0.04)c | 2.36E-06 | |
|
|
|||||
| TT (SE) | TA (SE) | AA(SE) | |||
|
|
|||||
| SNP04 | Total teat number | 12.60(0.17)a | 13.27(0.09)b | 13.58(0.06)c | 1.06E-07 |
| g.11,496A>T | Left teat number | 6.23(0.10)a | 6.62(0.05)b | 6.80(0.03)c | 3.92E-07 |
| [c.829A>T(S277C)] | Right teat number | 6.38(0.10)a | 6.64(0.06)b | 6.78(0.03)c | 0.00029 |
REFERENCES
Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265.


Bidanel JP, Rosendo A, Iannuccelli N, Riquet J, Gilbert H, Caritez JC, Billon Y, Amigues Y, Prunier A, Milan D. 2008. Detection of quantitative trait loci for teat number and female reproductive traits in Meishan×Large White F2 pigs. Animal 2:813–820.


Cassady JP, Johnson RK, Pomp D, Rohrer GA, Van Vleck LD, Spiegel EK, Gilson KM. 2001. Identification of quantitative trait loci affecting reproduction in pigs. J Anim Sci 79:623–633.


Cho IC, Park HB, Yoo CK, Lee GJ, Lim HT, Lee JB, Jung EJ, Ko MS, Lee JH, Jeon JT. 2011. QTL analysis of white blood cell, platelet and red blood cell-related traits in an F2 intercross between Landrace and Korean native pigs. Anim Genet 42:621–626.


Fuchs E, Haas AK, Spooner RA, Yoshimura S-i, Lord JM, Barr FA. 2007. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J Cell Biol 177:1133–1143.



Lewontin RC. 1964. The interaction of selection and linkage. I. General considerations; Heterotic models. Genetics 49:49–67.




Martínez-Giner M, Noguera JL, Balcells I, Alves E, Varona L, Pena RN. 2011. Expression study on the porcine PTHLH gene and its relationship with sow teat number. J Anim Breed Genet 128:344–353.


Oh HG, Jung WY, Yu SL, Lee JH. 2011. Expression analysis of porcine endogenous retroviruses (PERVs) in Korean native pig organs. CNU J Agric Sci 38:71–77.
Pumfrey RA, Johnson RK, Cunningham PJ, Zimmerman DR. 1980. Inheritance of teat number and its relationship to maternal traits in swine. J Anim Sci 50:1057–1060.


Seo KS, Kim SH, Park YI. 1996. Estimation of genetic parameters for teat numbers in swine. Korean J Anim Sci 38:133–138.
- TOOLS
-
METRICS

- Related articles
-
Effects of
CSN1S2 Genotypes on Economic Traits in Chinese Dairy Goats2013 July;26(7)





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




