 |
 |
| Anim Biosci > Volume 29(5); 2016 > Article |
|
Abstract
The objective of this study was to evaluate the present conventional selection program of a swine nucleus farm and compare it with a new selection strategy employing genomic enhanced breeding value (GEBV) as the selection criteria. The ZPLAN+ software was employed to calculate and compare the genetic gain, total cost, return and profit of each selection strategy. The first strategy reflected the current conventional breeding program, which was a progeny test system (CS). The second strategy was a selection scheme based strictly on genomic information (GS1). The third scenario was the same as GS1, but the selection by GEBV was further supplemented by the performance test (GS2). The last scenario was a mixture of genomic information and progeny tests (GS3). The results showed that the accuracy of the selection index of young boars of GS1 was 26% higher than that of CS. On the other hand, both GS2 and GS3 gave 31% higher accuracy than CS for young boars. The annual monetary genetic gain of GS1, GS2 and GS3 was 10%, 12%, and 11% higher, respectively, than that of CS. As expected, the discounted costs of genomic selection strategies were higher than those of CS. The costs of GS1, GS2 and GS3 were 35%, 73%, and 89% higher than those of CS, respectively, assuming a genotyping cost of $120. As a result, the discounted profit per animal of GS1 and GS2 was 8% and 2% higher, respectively, than that of CS while GS3 was 6% lower. Comparison among genomic breeding scenarios revealed that GS1 was more profitable than GS2 and GS3. The genomic selection schemes, especially GS1 and GS2, were clearly superior to the conventional scheme in terms of monetary genetic gain and profit.
Animal breeding schemes need to be well-designed to maximize long-term genetic gains obtained from genomic information (Henryon et al., 2014). To optimize the design of breeding programs, a full understanding of selection index theory is needed to predict the outcome of performance recording, genetic evaluation and subsequent selection (Toghiani, 2012). With advances in molecular technology or new biological tools, researchers have established methods to incorporate variations at the very basic DNA level into breeding programs. Genomic tools, such as single nucleotide polymorphism (SNP), have led to a new method known as “genomic selection.” This method, which was developed by Meuwissen (2007), utilizes dense SNP genotypes covering the entire genome to predict the breeding value. Genomic selection (GS) is currently being discussed as a potential method for optimization of breeding strategies for all livestock species (Thomasen et al., 2014). ZPLAN+ is a powerful tool that can evaluate and optimize breeding programs of large complexity (Täubert et al., 2010). ZPLAN+ integrates the possibility to directly include genomic information following the approach proposed by Dekkers (2007), with slight modifications, thereby enabling comparison of conventional and genomic breeding strategies. Accordingly, it is essential to assess the potential of using genomic information obtained via software such as ZPLAN+. The use of genomic information in swine breeding programs must be evaluated carefully before it is implemented, and economic aspects need to be considered. The main objective of this study was to optimize a swine breeding program with high genetic gains and low breeding cost. Additionally, conventional and genomic breeding programs are compared herein, and application of different GS strategies in swine breeding programs are investigated for their selection accuracy and expected selection responses.
The computer program, ZPLAN+ (Täubert et al., 2010), a user-friendly interface software, was used to simulate and evaluate different breeding schemes. This deterministic software allows modeling of all related breeding structures, as well as genetic and economic parameters, to account for complex breeding programs with a special emphasis on genomic information. ZPLAN+ was developed based on the gene flow method (Hill, 1974), selection index procedure for predicting reliabilities by Hazel and Lush (1942), and complex economic modeling. Breeding schemes were compared in terms of generation interval, monetary genetic gain, breeding costs, returns and discounted profit.
Four selection strategies were modeled in ZPLAN+ (Table 1). The first strategy reflected the current conventional breeding program, which was a progeny test system (CS). The second strategy was a scheme based strictly on genomic information (GS1). The third scenario (GS2) was created based on genomic information plus the pedigree and performance. The last scenario was a mixture of three scenarios (GS3), where selections were based on pedigree, genomic enhanced breeding value (GEBV) of the animals, performance and progeny information. In this study, we focus on the selection strategies of the sire line. Heritability, phenotypic standard deviations and economic weights of the breeding goal traits in this simulated breeding scheme are presented in Table 2. The breeding goal traits considered were the average daily gain (ADG), back fat thickness (BF), feed conversion rate (FCR), pH, meat color (L*) and intra-muscular fat (IMF). The phenotypic and genetic correlations among traits are shown in Table 3. The genetic parameter estimates used in this simulation study were obtained from National Swine Improvement Federation (NSIF, 1987) Guidelines for Uniform Swine Improvement Programs website. Economic values were based on the study of Haberland et al. (2014) and some were self-estimated. All cost figures are presented in US dollars.
This approach imitated the current breeding program, which was a progeny test system (CS). Genetic evaluation was based entirely on phenotypic records, pedigree and progeny information without considering any of the marker information. The breeding nucleus of purebred Duroc consisted of 300 sows that were mated naturally to 60 active boars, 35% of which were progeny tested. The genes are transferred to the production unit by a larger pool of 110 boars through artificial insemination, with the three-way crossbreeding scheme, terminal sire line is mated to F1 crossbred sows (Yorkshire×Landrace). The AI boars are assumed to be selected with a lower intensity than the breeding boars, but in which the breeding boars are included. In ZPLAN+, we split the boars into two selection groups, young boars (YB) and senior boars (SB). YB and SB united to be herd boars. The herd sows (HS) had only one selection group, which was the realization group of genetic gains generated by boars. Realization defines which group of animals are able to realize a defined performance of the breeding goal.
The traits that were included in field performance test were the ADG, BF, and FCR. Methods of measuring these traits were discussed by Cabling et al. (2015) in their study of genetic associations between production and meat quality traits. A total of 50 YB were selected in the first step of selection from 1,000 animals that entered the performance test. The selection criteria were based on their own performance on field test and pedigree. The cost for obtaining the pedigree was considered to be zero. The field performance test cost was $40 per male candidate, and it was completed at six months of age, before the candidates entered the two months acclimatization at the AI station. In the second selection step, 23 SB were selected and kept for another eight months based on their progeny records. Progeny testing was conducted by testing 3 purebred progeny on field test and 30 crossbreds on station test for meat quality traits such as pH, meat color (L*) and IMF. The production unit consisted of 15,000 crossbred sows, which was also the realization group of genetic gains generated by boars. The relative proportion of all inseminations was 30% use of YB and 70% use of SB. YB will only become sow sires, not sire of sons. This flow of genes was used in all selection schemes.
Three GS strategies were designed to identify the best approach for implementing GS in the swine breeding farm. The background for implementing genomic information in the selection index on this basis was developed by Dekkers (2007) and modified by Haberland et al. (2010) and Daetwyler et al. (2008; 2010). This approach requires correlation of the true breeding value and the GEBV to define every genomic trait rGBV. This was conducted using the approach developed by Erbe et al. (2011) based on an equation derived by Daetwyler et al. (2008):
where, N is the size of the reference population, and r2 is the reliability of the GEBV of the animals used in the reference population. In our calculations, we assumed Np = 1,000, which may be considered a minimum for GS in pigs and r2 was assumed to be 0.49 for all traits (Haberland et al., 2013). The proportion of genetic variance explained by markers (q2) was assumed to be 0.8 for all breeding goal traits as suggested by Erbe et al. (2011). Me was the number of independently segregating chromosome segments, which was derived by Goddard et al. (2011) as:
where, Ne denotes the effective population size, L is the average length of a chromosome in Morgans and k is the number of chromosome pairs. Assuming Ne = 100, k = 19 and L = 1.2 Morgans, the value of Me was 1,000. This assumption was based on a study conducted by Haberland et al. (2014). Thus, the accuracy of genomic breeding values was 0.577. Each selection scheme is discussed in detail below.
This scenario applied a one-step selection. Male candidates were genotyped in the early stage. The selection was based strictly on GEBV of the animal and pedigree. Here, 50 genotyped YB were selected from 1,000 candidates to enter the YB selection group. The top 23 YB comprised the SB group.
Male candidates were genotyped before they entered the field performance test. The test was conducted after six months. The basis of selection was strictly based on GEBV plus the performance test. Moreover, 50 genotyped YB were selected out of 1,000 candidates to enter the YB selection group. The top 23 YB were the SB group.
In this strategy, the selection steps were the same as CS except that male piglets were genotyped before they entered the field performance test. The male piglets were genotyped at 21 days of age. Out of 1,000 males genotyped, only 50 YB were selected based on the performance test and GEBV. The 50 YB were mated to breeding sows naturally for the progeny test and to produce the next generation of HS. Based on progenies information and owned GEBV, 23 SB were selected. SB were selected for the selection paths SB>HB and SB>HS.
The performance testing was assumed to cost an average of $55 per tested pig and $40 for station testing. The costs for were based on a study conducted by Akanno et al. (2013). For genomic breeding programs, the cost of genotyping was assumed to be $120 per pig based on the aforementioned study conducted by Akanno et al. (2013) and Haberland et al. (2014). The cost per selection strategy depends on the information source of each selection group. In all scenarios in ZPLAN+, the time unit was defined as half year and an investment period of ten years. The annual interest rate was assumed to be 5% for calculation of the discounted cost and 3% for calculation of the discounted return. The fixed costs in all selection schemes in this study were not accounted for due to the complexity of their determination and because only variable costs had an impact on the efficiency of the breeding strategy.
Four parameters influence genetic gain: selection intensity, accuracy of selection, genetic variance of the breeding goal and generation interval. Table 4 provides an overview of mean generation intervals and accuracies derived from a multiple trait selection index for all traits, including correlated information. ZPLAN+ calculates the generation interval directly from the gene flow-matrix. The mean generation interval in the reference scenario was 1.88 year. The generation intervals of GS1 and GS2 were reduced to 1.67 year. GS3 has the same generation interval with the reference scenario.
The accuracy of the selection index (rTI) for senior boars was as high as 0.91 in the conventional breeding scheme. This accuracy is higher than that of GS1 (0.68) and GS2 (0.70), assuming that the reference population size is 1,000. However, GS3 had a higher accuracy than CS (0.92). In terms of using YB, GS strategies have greater accuracy than conventional methods. GS1 is 26% higher than CS, while it was 31% higher for GS2 and GS3. Haberland et al. (2010) also concluded that YB can be selected more accurately by integrating genomic information based on the selection index. In their study, GS was found to be 47% higher than CS when N = 500. Furthermore, significantly higher accuracy observed for the genotyped YB in this study, which was analogous to the results reported by Tang et al. (2014) and Lillehammer et al. (2013). Although the reported increase differed in magnitude, these commonly increased in accuracy for the genotyped young males. Nevertheless, they were still not comparable to the levels observed for the conventionally selected senior males, which were based on sufficient progeny information.
The trend for the accuracy of selection index for boars depending on reference population size was also investigated (Figure 1). Further increases in accuracy for young and senior boars by enlarging the N have been discussed in many studies (Simianer, 2009; Li, 2014; Van Grevenhof and Van der Werf, 2015). The same trend was observed in the present study (Figure 1). Haberland et al. (2013) also investigated the significance of the size of the reference population and concluded that N = 1,000 could be considered a minimum size for a reference population in pig breeding.
The main goal of each breeding program was maximization of the genetic gain per generation and per year. The overall and individual trait monetary discounted genetic gains of the breeding goal per year are shown in Table 5. The CS breeding scheme improved the ADG by 19.868 g annually. In comparison with the GS scheme, GS1 can further improve that trait by 10%, while GS2 and GS3 increased this value by 18% and 8%, respectively. Furthermore, conducting CS would create an annual decrease of 0.36 mm in BF. Implementing GS using GS2 would decrease the BF by 0.41 mm which is 11% higher than CS. GS1 and GS3 had less BF reduction compare to CS. In the case of FCR, an annual reduction of 0.032 kg in feed intake was realized per kg of body mass gain. This reduction was enhanced by 7% (GS1), 15% (GS2), or 8% (GS3) when conducting GS. In terms of improving the meat quality traits, CS improved the pH by 0.027. By applying GS, this improvement increased by 17% when GS1 and GS3 was applied while 14% under GS2. Concurrently, the IMF increased by 0.301 under the CS scheme. This improvement increased by 11% when conducting GS3 while 8% and 12% using GS2 and GS3, respectively. Miar et al. (2014) found similar results and concluded that GS has great potential for the improvement of difficult to measure phenotypes and low heritable traits such as those influencing pork quality. In their study, they investigated the GS of pork pH in purebred pigs.
Applying the conventional selection strategy may create an annual monetary genetic gain (AMGG) of $5.80 (Table 5). When compared with GS strategies, GS1, GS2, and GS3 increased the AMGG by 10%, 12%, and 9%, respectively. These findings confirm the results of former studies that genetic gains were higher upon incorporation of genomic information (Schaeffer, 2006; Täubert et al., 2011).
The investment period carried out in this study was 10 years. The costs, returns and profit with relative percentage in each selection scheme are shown in Table 6. The fixed costs were not considered for every selection scheme in this study; thus, profits were higher than expected. As projected, the discounted costs of GS strategies were higher than those of CS. Specifically, the costs of GS1, GS2, and GS3 were 35%, 73%, and 89% higher than those of CS, respectively, assuming a genotyping cost of $120. Conducting CS may result in a discounted return per animal of $38.50. This return increases by 14% (GS1), 17% (GS2), or 13% (GS3) when adopting the genomic selection schemes, assuming 1,000 genotyped male candidates and a reference population size of 1,000. However, the relative profit of GS1 was 8% higher than that of CS, while it was only 2% higher under GS2 and 6% lower for GS3. In this study, as much as 8% extra income was possible when implementing GS. Furthermore, this extra income may further increase by reducing the genotyping cost. Several studies have been conducted to reduce the costs associated with genotyping (Ibañez-Escriche and Gonzalez-Recio, 2011). Abell et al. (2014) estimated that the genotyping cost per animal was $115 and $55 for high- and low-density panels, respectively. In Korea, the current genotyping cost per animal is around $300 (Seo, personal communication, 2015).
The possible use of high density (HD) and low density (LD) panels to reduce genotyping cost was also investigated in this study. Here, HD and LD were fitted in a two-stage selection. The selection steps were the same as GS3 but LD genotyping was carried out in first stage of selection of YB and HD genotyping was used in selecting SB. The cost of HD and LD genotyping were based on the study of Abell et al. (2014) but adjusted to be able to compare on the existing selection strategies. The genotyping cost per animal using LD is $60 while $120 for HD panels. The results showed that the use of HD and LD panels reduced the over-all cost by 21% and it gives 11% increase in profit compared to GS3. However, further study is needed on the difference of estimation of accuracy of GEBV between HD and LD panels and the possible effect on the over-all genetic gain.
GS1 was found to be the most profitable selection strategy, but had a lower accuracy than CS and GS3. GS2 produced the same results of GS1, but with lower profit. GS3 had much better accuracy of selection index, but was not profitable relative to CS. When GS2 is used, the traits to be tested must have high economic value and less cost so that commercial breeders will have greater confidence in the value of the selection index. The use of HD and LD panels reduced the genotyping cost. Overall, the results presented herein suggest that GS can optimize swine breeding programs, especially when the genotyping cost is reduced. However, implementation of GS in pig breeding remains a business decision to be made by individual breeding companies. Any possible extra income for the breeding company depends on whether customers are willing to pay more for improved genetic quality.
ACKNOWLEDGMENTS
This research was supported by Golden Seed Project, Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Service (KFS).
Figure 1
Trends in accuracy of selection index for senior boars depending on reference population size. CS, conventional system; GS, genomic selection.
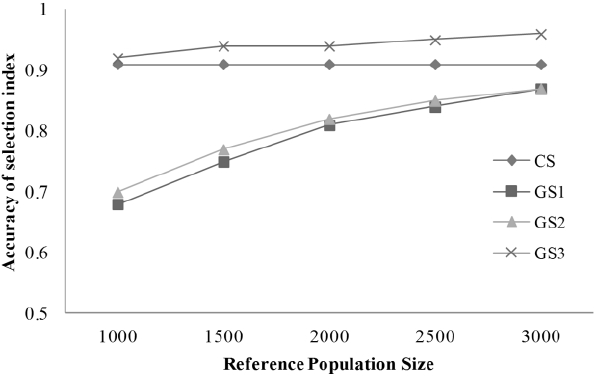
Table 1
Selection schemes modeled in ZPLAN+
| Scenario | Information sources |
|---|---|
| CS | Pedigree+Own performance+Progeny |
| GS1 | Pedigree+GEBV |
| GS2 | Pedigree+GEBV+Own performance |
| GS3 | Pedigree+GEBV+Own performance+Progeny |
Table 2
Heritability (h2), phenotypic standard deviation (σP) and economic weights (w) per unit of considered traits
| Trait | w ($) | σP | h2 |
|---|---|---|---|
| ADG (g/d) | 0.05 | 90.0 | 0.30 |
| BF (mm) | −15.0 | 5.08 | 0.52 |
| FCR (kg/kg) | −40.0 | 0.113 | 0.30 |
| pH | 20 | 0.25 | 0.38 |
| L* | - | 2.65 | 0.29 |
| IMF (%) | 9.25 | 1.0 | 0.47 |
Table 3
Phenotypic (above diagonal) and genotypic (below diagonal) correlations between considered traits
Table 4
Accuracy of selection index and mean generation intervals
| Scenario | CS | GS1 | GS2 | GS3 |
|---|---|---|---|---|
| Young boar | 0.54 | 0.68 | 0.70 | 0.70 |
| Senior boar | 0.91 | 0.68 | 0.70 | 0.92 |
| Mean generation interval | 1.88 | 1.67 | 1.67 | 1.88 |
Table 5
Discounted monetary genetic gain per year with relative percentage
REFERENCES
Abell C, Dekkers J, Rothschild M, Mabry J, Stalder K. 2014. Total cost estimation for implementing genome-enabled selection in a multi-level swine production system. Genet Sel Evol 46:32



Akanno EC, Schenkel FS, Friendship RM, Robinson JAB. 2013. Relative economic returns from selection schemes for a nucleus swine breeding program. Livest Res Rural Dev 25:Article #53
Cabling MM, Kang HS, Lopez BM, Jang M, Kim HS, Nam KC, Choi JG, Seo KS. 2015. Estimation of genetic associations between production and meat quality traits in Duroc pigs. Asian Australas J Anim Sci 28:1061–1065.




Daetwyler HD, Pong-Wong R, Villanueva B, Woolliams JA. 2010. The impact of genetic architecture on genome-wide evaluation methods. Genetics 185:1021–1031.



Daetwyler HD, Villanueva B, Woolliams JA. 2008. Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS ONE 3:e3395



Dekkers JCM. 2007. Prediction of response to marker-assisted and genomic selection using selection index theory. J Anim Breed Genet 124:331–341.


Erbe M, Reinhardt F, Simianer H. 2011. Empirical determination of the number of independent chromosome segments based on cross-validated data. In : 62nd Annual Meeting of the European Federation of Animal Science; Stavanger, Norway.
Goddard ME, Hayes BJ, Meuwissen THE. 2011. Using the genomic relationship matrix to predict the accuracy of genomic selection. J Anim Breed Genet 128:409–421.


Haberland AM, Luther H, Hofer A, Tholen E, Simianer H, Lind B, Baes C. 2014. Efficiency of different selection strategies against boar taint in pigs. Animal 8:11–19.


Haberland AM, Pimentel ECG, Ytournel F, Erbe M, Simianer H. 2013. Interplay between heritability, genetic correlation and economic weighting in a selection index with and without genomic information. J Anim Breed Genet 130:456–467.


Haberland AM, Ytournel FF, Luther H, Simianer H. 2010. Evaluation of selection strategies including genomic breeding values. In : Pigs 61st Annual Meeting of the European Association for Animal Production; Heraklion, Greece. p. 354
Henryon M, Berg P, Sørensen AC. 2014. Animal-breeding schemes using genomic information need breeding plans designed to maximise long-term genetic gains. Livest Sci 166:38–47.

Hill WG. 1974. Prediction and evaluation of response to selection with overlapping generations. Anim Sci 18:117–139.

Ibañez-Escriche N, Gonzalez-Recio O. 2011. Review. Promises, pitfalls and challenges of genomic selection in breeding programs. Spanish J Agric Res 9:404–413.

Li K. 2014. Modelling Genomic Selection Schemes in Bavarian Pig Breeding programs using ZPLAN+. PhD Thesis. Technische Universität München; München, Germany:
Lillehammer M, Meuwissen TH, Sonesson AK. 2013. Genomic selection for two traits in a maternal pig breeding scheme. J Anim Sci 91:3079–3087.


Meuwissen T. 2007. Genomic selection: marker assisted selection on a genome wide scale. J Anim Breed Genet 124:321–322.


Miar Y, Plastow G, Bruce H, Kemp R, Charagu P, Zhang C, Huisman A, Wang Z. 2014. Genomic selection of pork pH in purebred pigs for crossbred performance. In : 10th World Congress of Genetics Applied to Livestock Production; Vancouver, Canada.
NSIF. 1987. Guidelines for Uniform Swine Improvement Programs. Knoxville, TN, USA:
Schaeffer LR. 2006. Strategy for applying genome-wide selection in dairy cattle. J Anim Breed Genetics 123:218–223.

Simianer H. 2009. The potential of genomic selection to improve litter size in pig breeding programmes. In : 60th Annual Meeting of the European Association for Animal Production; Barcelona, Spain. p. 210
Tang G, Liu J, Xue J, Yang R, Liu T, Zeng Z, Jiang A, Jiang Y, Li M, Zhu L, Bai L, Shuai S, Wang Z, Li X. 2014. Optimizing selection strategies of genomic selection in swine breeding program based on a dataset simulated. Livest Sci 166:111–120.

Täubert H, Reinhardt F, Simianer H. 2010. ZPLAN+ A new software to evaluate and optimize animal breeding programs. In : Proceedings of the 9th World Congress on Genetics Applied to Livestock Production; Leipzig, Germany.
Täubert H, Rensing S, Reinhardt F. 2011. Comparing conventional and genomic breeding programs with ZPLAN+. Interbull Bull 162–168.
Thomasen JR, Egger-Danner C, Willam A, Guldbrandtsen B, Lund MS, Sorensen AC. 2014. Genomic selection strategies in a small dairy cattle population evaluated for genetic gain and profit. J Dairy Sci 97:458–470.


Toghiani S. 2012. Quantitative genetic application in the selection process for livestock production. Livestock Production, Dr. Khalid Javed edn. http://www.intechopen.com/books/livestock-production/quantitative-genetic-application-in-the-selectionprocess-for-livestock-productionAccessed October 3, 2015

- TOOLS





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





