 |
 |
|
|
Abstract
c-MYC (v-myelocytomatosis viral oncogene homologue) is a transcription factor that plays important role in many biological process including cell growth and differentiation, such as myogenesis and adipogenesis. In this study, we aimed to detect MYC gene polymorphisms, their genotype frequencies and to determine associations between these polymorphisms and meat quality traits in Berkshire pigs. We identified a single nucleotide polymorphism (SNP) in intron 2 of MYC gene by Sanger sequencing, i.e., g.3350G>C (rs321898326), that is only found in Berkshire pigs, but not in other breeds including Duroc, Landrace, and Yorkshire pigs that were used in this study. Genotypes of total 378 Berkshire pigs (138 sows and 240 boars) were determined using Hha I restriction enzyme digestion after polymerase chain reaction. Observed allele frequencies of GG, GC, and CC genotypes were 0.399, 0.508, and 0.093 respectively. Statistical analysis indicated that the g.3350G>C polymorphism was significantly associated with pH45min and cooking loss (p<0.05), suggesting that g.3350G>C SNP can be used for pre-selection of pH45min and cooking loss traits in Berkshire pigs.
Swine industry has mostly focused on improving productivity of pork in respect to quantity and distribution of fresh meat, which has entailed a decline in quality of pork loin (Cameron, 1990; Cliplef and McKay, 1993). In Korea, fresh pork is in great demand as it is widely consumed roasted or boiled. Basically, consumer perception of meat quality is an important selection criterion for the purchase of meat. However, varying degrees in meat quality decreases reliability and challenges consumers in making good pork purchases (Jung et al., 2011). Therefore, to overcome this problem, industries have developed a system to improve traits accounting for meat quality to satisfy the consumer demand for the highest quality of pork (Lee et al., 2012).
Among various pig breeds, color breeds are typically known to have higher marbling scores than white breeds. In Japan, the price of Berkshire meat is 50% higher than that of regular commercial pork (Crawford et al., 2010). In Korea, Berkshire pork is also sold as premium meat in the market (Do et al., 2012). The preference for black pig meat is partly attributed to consumers’ increasing demand for high quality pork. However, insufficient supply is an obstacle to increased consumption at present. Such rise in consumer expectation has ultimately triggered vigorous genetics-based research in Berkshires to improve breeding and management skills for enhanced pork productivity and quality (Jeong et al., 2010; Lee et al., 2010; Park et al., 2010; Kang et al., 2011).
Porcine v-myelocytomatosis viral oncogene homologue (MYC) gene, encoding the c-MYC protein, consists of 3 exons and 2,464 base pairs and is located on chromosome 4. The c-MYC protein is considered as an important transcriptional regulatory factor during cytogenesis and the gene expression of MYC gene is reported to be associated with several physiological processes that involve hormones, growth factors, cytokines, lymphokines, nutritional status, development, differentiation etc (Levens et al., 1997). Also, its function has been associated with myogenesis (Miner and Wold, 1991; Whitelaw and Hesketh, 1992), muscle hyperplasia (Kipshidze et al., 2002), and adipogenesis (Freytag and Geddes, 1992; Ninomiya-Tsuji et al., 1993; MacDougald and Lane, 1995). Our previous study indicated that a non-synonymous SNP of porcine MYC gene might be associated with a few economic traits such as age at 90 kg. However, to our knowledge there is no previous study that has studied the relationship between porcine MYC gene polymporphism and meat quality traits in pigs. Therefore, in this study, we identified a Berkshire breed specific single nucleotide polymporphism and investigated the polymorphism of c-MYC gene with meat quality traits in Berkshire pigs
First, to detect breed-specific variations in the MYC gene, 20 blood samples from each of four types of purebred pigs in Korea – Berkshire, Duroc, Landrace, and Yorkshire – were collected in anticoagulant containing vacutainers. Genomic DNA was isolated from the blood samples using the G-DEX IIb Genomic DNA Extraction Kit (IntronBio Inc., Seoul, Korea).
To measure meat quality traits studied in this study, muscle samples were obtained from 378 Berkshire carcasses (138 sows, 240 boars) shipped from a slaughter house located in Namwon city of Chonbuk province, Republic of Korea. Genomic DNA was extracted from muscle samples using the same extraction kit mentioned above. All procedures were followed according to the manufacturer’s instructions.
The DNA sequence of porcine MYC gene was retrieved from NCBI database (accession number: NC_010446.4). As shown in Table 1, Primer 3 program (http://frodo.wi.mit.edu/primer3/) was used to design 8 primer sets (P1-P8) for full length sequencing of the MYC gene by polymerase chain reaction (PCR).
The PCR reaction mixture was composed of 1.5 μL of genomic DNA (25 ng), 0.2 pmole of forward and reverse primers, 200 nM of dNTP, 2. 5 unit of Taq polymerase, and 1× reaction buffer of 10 mM of Tris-HCl (pH 9.0), 50 mM of KCl, 1.4 mM of MgCl2 and 1% of Triton X-100 with a total volume of 20 μL. The thermal cycling condition of the mixture included denaturation at 94ºC for 5 min, and 35 cycles of PCR amplification conducted for 30 s at different annealing temperatures for each primer set (Table 1) with primer extension at 72ºC for 40 s. Final extension was performed at 72ºC for 5 min. Electrophoresis of the amplified products was conducted on 1.5% agarose gels stained with ethidium bromide (EtBr).
Twenty DNA samples pooled from each of the four breeds (Berkshire, Duroc, Landrace, and Yorkshire) were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kits (Applied Biosystems, Foster City, CA, USA) and the ABI 3130 Genetic Analyzer (Applied Biosystems, USA). The genotyped sequences were subject to peak and alignment analyses using the SeqMan 7.0 program (DNASTAR, Madison, WI, USA) to detect polymorphism in the gene and determine breed-specificity.
To determine genotypes from Berkshire-specific polymorphism that was detected in the earlier step (Table 2), polymerase chain reaction-restriction length polymorphism (PCR-RFLP) analysis was performed on genomic DNA from muscle samples amplified by PCR. The amplified products of PCR (2 μL) were treated with 1 unit of HhaI restriction enzyme, 1 μL of 10× restriction enzyme buffer and sterilized distilled water, giving a total volume of 10 μL. After digestion reactions took place for restriction enzyme-specific temperature and time, the DNA fragments were separated on 1.5% agarose gel in 1× TAE buffer containing EtBr for genotyping.
After slaughter, a section of loin at the 10th rib of the carcass was taken to assess meat quality based on the four meat quality traits – pH/color, drip loss, cooking loss, and shear force.
Change in pH and color of carcass at 45 min and 24 h postmortem were measured with a pH meter (Model 720, Thermo Orion, Pittsburgh, PA, USA) and a Chromameter (CR-300, Minolta Camera Co, Osaka, Japan) respectively. The Commission Internationale de l’Eclairage (CIE) L* values were read from the measurement.
A section of size-standardized loin was cut from 24 h postmortem carcass and weighed. After measuring its weight, the samples were suspended in inflated polyethylene bags and stored at 2ºC for 48 h and reweighed. Drip loss was calculated and expressed as the percent change in weight.
After cutting size-standardized section of loin samples from 24 h postmortem carcass, the samples were sealed in suspended polyethylene bags and immersed in a heated water bath until internal temperature was 72ºC. Cooking loss was measured and expressed as the percentage of initial sample weight.
Upon heating the pork loin at 72ºC, a cylindrical shearing device (1.27 cm in diameter) was used to collect samples (parallel to muscle fiber orientation). Following the sample collection, a measuring tool (SeriesIX, Instron Corp, Norwood, MA, USA) was used to measure the maximum force (N) required to cut the cylindrical samples (perpendicular to the long axis).
SAS 9.1 (SAS, USA) program was used to estimate the genotype effect of MYC gene on meat quality traits by general linear model procedure. The least significance difference test was implemented to determine significance between mean values. The model used in this study to estimate the genotype effect on the traits is as follows:
Where,
Porcine MYC sequences were retrieved from the NCBI database (accession number: NC_0104464.4). The MYC gene is located on chromosome 4 comprising three exons and 2,464 base pairs with coding regions of 1,359 bp, and 5′ untranslated region (UTR) and 3′ UTR at lengths of 591 bp and 514 bp respectively. Full length sequencing of MYC gene was performed for pooled DNAs from each four pig breeds (Berkshire, Duroc, Landrace, Yorkshire) commonly bred in Korea by using eight primer sets (P1-P8; Table 1). When compared with the MYC SNPs deposited in dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?geneId=448810), sequencing results showed that the g.3350G>C polymorphism in the intron 2 of porcine MYC gene was found only in Berkshire pigs, but not in other breeds (Duroc, Landrace, and Yorkshire, Table 2).
Once the polymorphic site (g.3350G>C) was detected in the MYC gene and identified to be Berkshire-specific, genotyping was carried out using the genomic DNA of muscle samples of Berkshire. Genotypes were determined with PCR-RFLP using HhaI restriction enzyme (Table 2). Digestion of PCR product with HhaI restriction enzyme yields two fragments with 480 bp and 201 bp respectively for allelotype G, whereas for allelotype C one fragment with 681 bp is produced (Figure 1). Allele frequencies of GG, GC, and CC genotypes of 378 Berkshire pigs were 0.399, 0.508, and 0.093 respectively (Table 3).
The meat quality measurements obtained from 378 Berkshire pigs in this study are shown in Table 4. Although the muscle samples used for this study were obtained from 138 sows and 240 boars, a significant difference between sexes was not observed across the meat quality traits. The pH of muscle, which affects freshness of meat due to postmortem metabolism, was 6.07±0.30 at 45 min and 5.79±0.22 at 24 h postmortem respectively. Meat color which was evaluated for lightness (L*), redness (a*), and yellowness (b*), was based on CIE LAB. Lightness (L*) was 50.49±3.00, redness (a*) was 16.36±1.16, and yellowness was 5.28±1.19 respectively. As for the water-holding capacity of meat, drip loss was 2.51±2.20 and cooking loss was 16.79±3.99. Shear force was 2.66±0.69. Based on the results concerning meat quality traits, the g.3350G>C SNP effect was significant for pH45min and cooking loss (Table 5, p<0.05). Berkshire pigs with the homozygous genotype C had the highest pH45min and smallest cooking loss (Table 5). Except two traits, the g.3350G>C SNP had no effects on other trait including pH24 h, meat colors and drip loss, and shear force (Table 5, p>0.05). There are several physical and chemical factors known to affect pork quality (Stalder et al., 1998). A previous study reported that postmortem pH in muscle and meat quality characteristics correlated (Huff-Lonergan et al., 2002). According to Jung et al. (2011), pH45 min and pH24 h were positively correlated whereas both were negatively correlated with drip loss and shear force in Berkshire pigs.
Porcine MYC gene was mapped on SSC4 (Reiner et al., 1998; Cepica et al., 2003), where particularly major quantitative trait locus regions for backfat at tenth rib (Liu et al., 2007; Edwards et al., 2008) and average daily gain (de Koning et al., 2001) reside. Porcine chromosome SSC 4p13 contains a synteny group, with F13B, ATP1B1, GBA, ATP1A1, IVL, and two microsatellites S0001, S0067 (Andersson et al., 1994; Marklund et al., 1999).
Given the fact that MYC plays a role in the cellular processes as a molecular hub that receives multiple signals and integates them, either negatively or positively, to control other sets of genes. Thus, MYC has not only as a single function, but also a multitude of different categories of cellular functions (Potter and Marcu, 1997). In this regard, MYC expression that is regulated by external cellular stimulus including hormones, growth factors, cytokines, lymphokines, nutritional status, development and differentiation (Levens et al., 1997), is connected to various physiological processes. More specifically, MYC is involved in myogenesis (Miner and Wold, 1991; Whitelaw and Hesketh, 1992), muscle hyperplasia (Kipshidze et al., 2002) and adipogenesis (Ninomiya-Tsuji et al., 1993; MacDougald and Lane 1995), hence the proto-oncogene could be a candidate gene contributing to variability of fatness in pigs. In addition, previous study has suggested the influence of MYC gene expression on glucose metabolism (Collier et al., 2003), whereby lactic acid production from glycolysis reaction might have an effect on the pH of meat.
Significant difference was also noted between GG, CG, and CC genotypes at pH45min, with measurements of 5.96±0.04, 5.97±0.04, and 6.13±0.07 respectively (Table 5). Muscle pH is an important economic factor that is associated with water-holding capacity of muscle’s, which in turn has a marked influence on the appearance of meat (Barge et al., 1991). According to Jung et al. (2011), pH45min and pH24min were positively correlated whereas both were negatively correlated with drip loss and shear force in Berkshire pigs. Previous studies have suggested the influence of MYC gene on glucose metabolism (Collier et al., 2003), whereby lactic acid production from glycolysis reaction has an effect on the pH of meat. Because pH45min was significantly higher in the animals with CC genotype of g.3350G>C polymorphism than the levels in the animals having other genotypes in this study, it is suggested that this genotype may play an influential role in improving pork quality.
Muscle structure could (or may) be affected by heat technique, composition of meat, and degree of heating meat. Heating of meat contracts muscle fibers that can cause decrease in water-holding capacity and cooking loss irrespective to the method of heating (Cho et al., 2008). High cooking loss is associated with poor meat quality (Schmidt et al., 2010) and is reported that the higher the marbling score is, the lower the cooking loss (Kim and Lee, 2003). Therefore, low cooking loss is regarded as a favorable trait of meat quality. Significant difference in genetic effect of GG, CG, and CC genotypes was identified with respect to loss, with measurements 16.38±0.63, 15.48±0.84, and 15.50±0.67 respectively (Table 5).
The current study detected Berkshire-specific polymorphism in porcine MYC gene (g.3350G>C). The polymorphic site was significantly associated with pH and cooking loss, which are known to have high economic value due to their influence on the characteristic of meat quality. The polymorphism detected from this study may be of considerable value as baseline data for research grounded in molecular genetics to improve meat quality of Berkshire breed. It is anticipated that further studies will lead to improved breeding and management strategies directed toward the production of higher quality meat in the future.
ACKNOWLEDGMENTS
This work was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ01110901), Rural Development Administration, Republic of Korea. The authors sincerely thank Dr. H. C. Park of Dasan Breeding Inc. for Berkshire muscle samples and phenotype data, and Ms. S. H. Moon for English editing.
Figure 1
Genomic structure and polymorphism of the MYC gene in Berkshire. Coding exons are marked by shaded blocks and 5′ and 3′ UTR by white blocks. (A) agarose gel displaying a HhaI restriction digest on an amplified portion of c.3350 G>C. M: 100 bp ladder; GG: 480 bp and 201 bp; GC: 681 bp, 480 bp and 201 bp; CC: 681 bp.
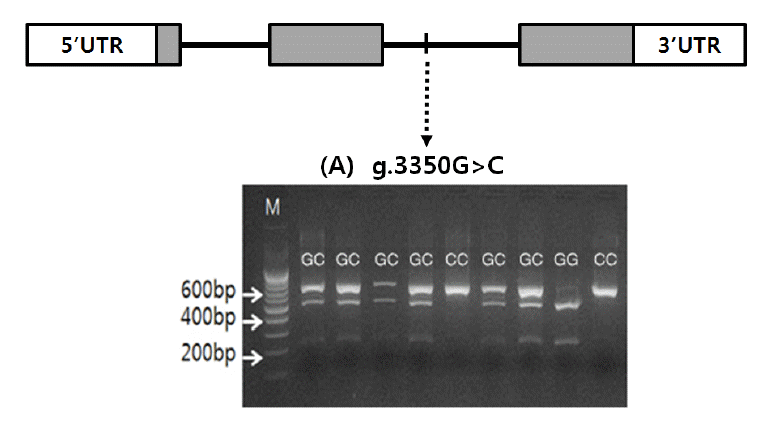
Table 1
Primer sets for sequencing of porcine MYC gene
Table 2
Information of PCR-RFLP for polymorphic site of c-MYC gene in Berkshire
| Primer set | Region | Position | Polymorphic site | Restriction enzyme | SNP IDa |
|---|---|---|---|---|---|
| RP1 | Intron 2 | 3350 | G>C | HhaI | rs321898326 |
PCR-RFLP, polymerase chain reaction-restriction length polymorphism; c-MYC, v-myelocytomatosis viral oncogene homologue; SNP, single nucleotide polymorphism.
a SNP ID is followed by dbSNP (http://www.ncbi.nlm.nih.gov/snp).
Table 3
Genotype and allele frequencies of polymorphic site of MYC gene in Berkshire
| Polymorphic site | Genotype | No. of animals | HOB | Alleles | Allele frequency | HWE (x2, p-value) | |
|---|---|---|---|---|---|---|---|
| g.3350G>C | GG | 151 | 0.399 | 0.427 | G | 0.653 | |
| GC | 192 | 0.508 | 0.453 | 0.018 | |||
| CC | 35 | 0.093 | 0.120 | C | 0.347 |
Table 4
Means and standard deviations for meat quality measurements in Berkshire
Table 5
Association analyses between the polymorphisms and individual traits in Berkshire
| Traits | g.3350G>C | ||
|---|---|---|---|
|
|
|||
| GG | GC | CC | |
| Muscle pH45min | 5.96±0.04b | 5.97±0.04b | 6.13±0.07a |
| Muscle pH24h | 5.79±0.03 | 5.78±0.03 | 5.79±0.05 |
| Lightness (L*) | 50.02±0.34 | 50.10±0.48 | 49.61±0.72 |
| Radness (a*) | 16.17±0.18 | 16.05±0.18 | 16.06±0.29 |
| Yellowness (b*) | 4.86±0.19 | 4.85±0.19 | 5.10±0.29 |
| Drip loss (%) | 3.46±0.35 | 3.33±0.35 | 3.56±0.52 |
| Cooking loss (kg) | 16.38±0.63b | 15.48±0.84b | 15.50±0.67a |
| Shear force (N) | 2.65±0.11 | 2.56±0.11 | 2.89±0.16 |
REFERENCES
Andersson L, Haley CS, Ellegren H, Knott SA, Johansson M, Andersson K, Andersson-Eklund L, Edfors-Lilja I, Fredholm M, Hansson I, Hakansson J, Lundstrøm K. 1994. Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science 263:1771–1774.


Barge MT, Destefanis G, Toscano GP, Brugiapaglia A. 1991. Two reading techniques of the filter paper press method for measuring meat water-holding capacity. Meat Sci 29:183–199.


Cameron ND. 1990. Genetic and phenotypic parameters for carcass traits, meat and eating quality traits in pigs. Livest Prod Sci 26:119–135.

Cepica S, Stratil A, Kopecny M, Blazkova P, Schroffel J, Davoli R, Fontanesi L, Reiner G, Bartenshlager H, Moser G. 2003. Linkage and QTL mapping for Sus scrofa chromosome 4. J Anim Breed Genet 120:28–37.

Cho SH, Kim JH, Seong PN, Cho YM, Chung WT, Park BY, Chung MO, Kim DH, Lee JM, Ahn CN. 2008. Physico-chemical meat quality properties and nutritional composition of hanwoo steer beef with 1++ quality grade. Korean J Food Sci An 28:422–430.

Cliplef RL, McKay RM. 1993. Carcass quality characteristics of swine selected for reduced backfat thickness and increased growth rate. Can J Anim Sci 73:483–494.

Collier JJ, Doan TT, Daniels MC, Schurr JR, Kolls JK, Scott DK. 2003. c-Myc is required for the glucose-mediated induction of metabolic enzyme genes. J Biol Chem 278:6588–6595.


Crawford SM, Moeller SJ, Zerby HM, Irvin KM, Kuber PS, Velleman SG, Leeds TD. 2010. Effects of cooked temperature on pork tenderness and relationships among muscle physiology and pork quality traits in loins from Landrace and Berkshire swine. Meat Sci 84:607–612.


Do CH, Cho BW, Lee DH. 2012. Study on the prolactin receptor 3 (PRL3) gene and the retinol-binding protein 4 (RBP4) gene as candidate genes for production traits in Berkshire pigs. Asian Australas J Anim Sci 25:183–188.



de Koning DJ, Rattink AP, Harlizius B, Groenen MAM, Brascamp EW, van Arendonk JAM. 2001. Detection and characterization of quantitative trait loci for growth and reproduction traits in pigs. Livest Prod Sci 72:185–198.

Edwards DB, Ernst CW, Tempelman RJ, Rosa GJM, Raney NE, Hoge MD, Bates RO. 2008. Quantitative trait loci mapping in an F2 Duroc×Pietrain resource population: I. Growth traits. J Anim Sci 86:241–253.


Freytag SO, Geddes TJ. 1992. Reciprocal regulation of adipogenesis by Myc and C/EBP alpha. Science 256:379–382.


Huff-Lonergan E, Baas TJ, Malek M, Dekkers JC, Prusa K, Rothschild MF. 2002. Correlations among selected pork quality traits. J Anim Sci 80:617–27.


Jeong DW, Choi YM, Lee SH, Choe JH, Hong KC, Park HC, Kim BC. 2010. Correlations of trained panel sensory values of cooked pork with fatty acid composition, muscle fiber type, and pork quality characteristics in Berkshire pigs. Meat Sci 86:607–615.


Jung JH, Kim CW, Park BY, Choi JS, Park HC. 2011. Genetic parameter estimates for meat quality traits in berkshire pigs. J Anim Sci Technol 53:289–296.

Kang YK, Choi YM, Lee SH, Choe JH, Hong KC, Kim BC. 2011. Effects of myosin heavy chain isoforms on meat quality, fatty acid composition, and sensory evaluation in Berkshire pigs. Meat Sci 89:384–389.


Kim CJ, Lee ES. 2003. Effects of quality grade on the chemical, physical and sensory characteristics of Hanwoo (Korean native cattle) beef. Meat Sci 63:397–405.


Kipshidze N, Iversen P, Keane E, Stein D, Chawla P, Skrinska V, Shankar LR, Mehran R, Chekanov V, Dangas G, Komorowski R, Haudenschild C, Khanna A, Leon M, Keelan MH, Mose J. 2002. Complete vascular healing and sustained suppression of neointimal thickening after local delivery of advanced c-myc antisense at six months follow-up in a rabbit balloon injury model. Cardiovasc Radiat Med 3:26–30.


Lee EA, Kim JM, Lim KS, Ryu YC, Jeon WM, Hong KC. 2012. Effects of variation in porcine MYOD1 gene on muscle fiber characteristics, lean meat production, and meat quality traits. Meat Sci 92:36–43.


Lee SH, Choi YM, Choe JH, Kim JM, Hong KC, Park HC, Kim BC. 2010. Association between polymorphisms of the heart fatty acid binding protein gene and intramuscular fat content, fatty acid composition, and meat quality in Berkshire breed. Meat Sci 86:794–800.


Levens D, Duncan RC, Tomonaga T, Michelotti GA, Collins I, Davis-Smyth T, Zheng T, Michelotti EF. 1997. DNA conformation, topology, and the regulation of c-myc expression. Curr Top Microbiol Immunol 224:33–46.


Liu G, Jennen DG, Tholen E, Juengst H, Kleinwächter T, Hölker M, Tesfaye D, Un G, Schreinemachers HJ, Murani E, Ponsuksili S, Kim JJ, Schellander K, Wimmers K. 2007. A genome scan reveals QTL for growth, fatness, leanness and meat quality in a Duroc-Pietrain resource population. Anim Genet 38:241–252.


MacDougald OA, Lane MD. 1995. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem 64:345–373.


Marklund L, Nystrøm PE, Stern S, Andersson-Eklund L, Andersson L. 1999. Confirmed quantitative trait loci for fatness and growth on pig chromosome 4. Heredity 82:134–141.


Miner JH, Wold BJ. 1991. c-myc inhibition of MyoD and myogenin-initiated myogenic differentiation. Mol Cell Biol 11:2842–2851.



Ninomiya-Tsuji J, Torti FM, Ringold GM. 1993. Tumor necrosis factor-induced c-myc expression in the absence of mitogenesis is associated with inhibition of adipocyte differentiation. Proc Natl Acad Sci USA 90:9611–9615.



Oh JD, Song KD, Seo JH, Kim DK, Kim SH, Seo KS, Lim HT, Lee JB, Park HC, Ryu YC, Kang MS, Cho S, Kim ES, Choe HS, Kong HS, Lee HK. 2014. Genetic traceability of black pig meats using microsatellite markers. Asian Australas J Anim Sci 27:926–931.




Park BY, Cho SH, Kim JH, Seong PN, Kang GH, Jeong DW, Kim C, Park HC, Jeong JH, Choi JS, Kim DH. 2010. Comparison of pork quality by different berkshire line. Korean J Food Sci Ani Resour 30:867–871.

Potter M, Marcu KB. 1997. The c-myc story: Where we’ve been, where we seem to be going. Curr Top Microbiol Immunol 224:1–17.

Reiner G, Hecht W, Leeb T, Brenig B, Robic A, Dzapo V. 1999. Isolation and characterization of the porcine c-myc proto-oncogene and chromosomal assignment to SSC 4p13. Anim Genet 30:204–206.


Schmidt TB, Schilling MW, Behrends JM, Battula V, Jackson V, Sekhon RK, Lawrence TE. 2010. Use of cluster analysis and preference mapping to evaluate consumer acceptability of choice and select bovine M. longissimus lumborum steaks cooked to various end-point temperatures. Meat Sci 84:46–53.







 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





