 |
 |
| Anim Biosci > Volume 29(2); 2016 > Article |
|
Abstract
A study was conducted to assess the genetic diversity among Simmental Cross cattle in West Sumatra using microsatellite DNA markers. A total of 176 individual cattle blood samples was used for obtaining DNA samples. Twelve primers of microsatellite loci as recommended by FAO were used to identify the genetic diversity of the Simmental Cross cattle population. Multiplex DNA fragment analysis method was used for allele identification. All the microsatellite loci in this study were highly polymorphic and all of the identified alleles were able to classify the cattle population into several groups based on their genetic distance. The heterozygosity values of microsatellite loci in this study ranged from 0.556 to 0.782. The polymorphism information content (PIC) value of the 12 observed loci is high (PIC>0.5). The highest PIC value in the Simmental cattle population was 0.893 (locus TGLA53), while the lowest value was 0.529 (locus BM1818). Based on the genetic distance value, the subpopulation of the Simmental Cross-Agam and the Simmental Cross-Limapuluh Kota was exceptionally close to the Simmental Purebred thus indicating that a grading-up process has taken place with the Simmental Purebred. In view of the advantages possessed by the Simmental Cross cattle and the evaluation of the genetic diversity results, a number of subpopulations in this study can be considered as the initial (base) population for the Simmental Cross cattle breeding programs in West Sumatra, Indonesia.
The Simmental Cross cattle in West Sumatra mostly originate from the crossing program between the Ongole Grade (in Indonesia known as Peranakan Ongole [PO]) and the Simmental Purebred sires. The superior properties of the Simmental Cross cattle are its good level of adaptability to tropical climates and strong ability to grow. As a result, the farmers in West Sumatra prefer using the Simmental Purebred semen (Siregar et al., 1999) for their cows when applying artificial insemination (AI) technology from Lembang Artificial Insemination Center (BIB Lembang) or Tuah Sakato Artificial Insemination Center (BIBD Tuah Sakato). In West Sumatra, artificial insemination has intensively used the Simmental Purebred semen for years resulting in an increased Simmental Cross cattle population in West Sumatra and has generated cattle populations with improved morphological profiles compared to the local cattle (Agung et al., 2014). This phenomenon inspired the farmers and the local Government to design a breeding program for the Simmental Cross cattle in West Sumatra.
The success of the breeding program can be affected by several factors, including determining the number of the initial (base) population in order to produce offspring consistent with the objectives of the breeding program. Recently, local farmers and the local Government of West Sumatra have been facing difficulties in determining the initial (base) population for the breeding program of the Simmental Cross cattle. This problem can be caused by several factors including insufficiency of genetic information about the present status of the Simmental Cross cattle, the lack of studies about genetic diversity in Simmental Cross cattle, and also the distribution of the Simmental Cross cattle in a large area of West Sumatra, Indonesia.
Microsatellites are almost ideal genetic markers because they are abundant, codominant, highly polymorphic, and are spread out across the entire euchromatic part of the genome (Bennett, 2000). Microsatellites can be used for estimating the genetic distance (Rehman and Khan, 2009), the relationship among livestock breeds (Maretto et al., 2012), and also the genetic diversity (Mao et al., 2008). This study was conducted to study the genetic diversity in the Simmental Cross cattle in West Sumatra using microsatellite markers as scientific evidence for the latest status of the Simmental Cross cattle in West Sumatra and also grouping the Simmental Cross cattle population in West Sumatra that can be used as the initial (base) population for breeding programs.
Blood samples were collected from herds owned privately and by artificial insemination centers. Genomic DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). A total of 176 DNA samples were collected and classified into three categories, i.e. Simmental Purebred, Simmental Cross, and Ongole Grade (PO) cattle (Table 2). Blood samples of Simmental Cross cattle were obtained from West Sumatera provinces and Karya Anugerah Rumpin (KAR) Farm in West Java, Indonesia. Blood samples of Simmental Purebred cattle were obtained from two locations, i.e. Lembang Artificial Insemination Center (BIB Lembang) in West Java and Tuah Sakato Artificial Insemination Center (BIBD Tuah Sakato) in West Sumatra, whereas the blood samples from Ongole Grade (PO) cattle were obtained from the Research Center for Biotechnology (RC Biotech.) Farm in West Java.
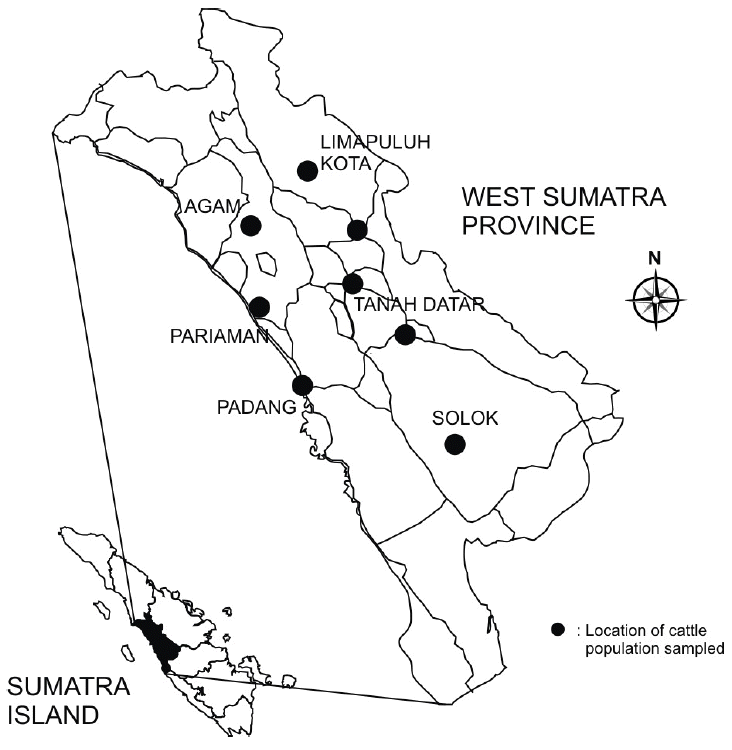
The sampling of the Simmental Cross cattle blood in West Sumatra in this study was divided into three areas based on the size of the Simmental cattle population i.e. high population (Limapuluh Kota), medium population (Agam, Padang, Payakumbuh, Solok, Sawahlunto, and Tanah Datar), and low population (Pariaman). The samples from Pariaman (an area with very low population of Simmental cattle) was included in the analysis as a comparison study and also to see the status of Simmental Cross cattle particularly in areas with very low population of Simmental cattle. Information about the Simmental cattle population in West Sumatra province was obtained from the Indonesian cattle population survey (Statistics Indonesia, 2011). Geographical distribution of the cattle sampled in West Sumatra is shown in Figure 1.
A total of 12 microsatellite labelled primers, which was recommended by FAO, were used in the polymerase chain reaction (PCR) process (primers sequence, annealing temperature, range of PCR product size, and label used are shown in Table 1). The PCR reagent composition is as follows: KAPA2G Robust HotStart ReadyMix PCR Kit (Kapa Biosystems, Cape Town, South Africa) (18 μL), forward and reverse labelled primers (200 ng/μL), nuclease free water, and DNA samples (5–30 ng/μL). The program in the PCR machine (Eppendorf, Hamburg, Germany) was set as follows: 94°C; 5 min (1 cycle), 35 cycles consisting of three stages: i) 94°C; 30 s, ii) 51°C to 59°C; 30 s (depends on primers), and iii) 72°C; 30 s, followed by 1 cycle at 72°C; 5 min. The PCR products were then visualized by electrophoresis using 2% agarose gel and followed by ethidium bromide staining. Multiplex DNA fragment analysis was afterwards used for allele identification. The multiplex DNA fragment analysis was conducted in 1st BASE Laboratory, Malaysia.
Result of the multiplex DNA fragment analysis was processed using CONVERT ver. 1.3.1 (Glaubitz, 2004), CERVUS ver. 3.0.7 (Kalinowski et al., 2007), MEGA ver. 6.0 (Tamura et al., 2013), and POPGENE ver. 1:32 (Yeh and Boyle, 1997) programs. The CONVERT program was used for conversion of the length of alleles which were observed for each individual sample to assure suitability for further analysis by POPGENE and CERVUS program. Data of the heterozygosity, the genetic distance, and the polymorphism information content (PIC) value were obtained from POPGENE ver. 1:32 and CERVUS ver. 3.0.7 analysis result. The genetic distance value was used to make a dendrogram that illustrates the relationship among cattle populations using MEGA ver. 6.0.
Bayesian clustering assignments were analysed by STRUCTURE ver. 2.2 (Pritchard et al., 2000). Ten independent runs were performed for each K between 2 and 10, with a burn-in period of 10,000 iterations followed by 100,000 iterations of the Markov Chain Monte Carlo algorithm. The STRUCTURE HARVESTER (Earl and vonHoldt, 2012), which implements the Evanno method (Evanno et al., 2005) was used to identify the optimal groups (K). The principal component analysis (PCA) and the discriminant analysis of principal components (DAPC) were carried out on the microsatellite genotypes to determine breed relationships. PCA and DAPC were performed using adegenet package (Jombart, 2008) of R ver. 3.2.0 (2015.4.16) (R Development Core Team, 2015).
Twelve microsatellite loci from the entire population were analysed and 317 alleles consisting of 83 alleles were detected in the Simmental Purebred, 143 alleles in the Simmental Cross, and 91 alleles in the Ongole Grade (PO). The 12 microsatellite loci revealed high polymorphism to evaluate the genetic diversity especially in the Simmental Cross population. The observed heterozygosity value ranged from 0.556±0.071 (KAR Farm) to 0.782±0.029 (Tanah Datar) and the expected heterozygosity value ranged from 0.605±0.061 (Pariaman) to 0.760±0.020 (LIPI Farm). The expected heterozygosity was higher than the observed heterozygosity in Limapuluh Kota, Agam, KAR Farm, Artificial Insemination Center, and LIPI Farm subpopulations (Table 2). This condition can be explained by several factors, including null alleles, assortative mating, the Wahlund effect, selection against heterozygotes, inbreeding, or a combination of all these factors (Cervini et al., 2006). In addition, the low value of heterozygosity indicates that certain breeds are relatively well-conserved (Czerneková et al., 2006).
In this study, the locus TGLA53 allele 168 is a private allele for the Simmental Purebred. This allele is a specific allele candidate for the Simmental Purebred cattle population and needs to be validated by further studies. Information about the private allele for the Simmental cattle was limited. Putnova et al. (2011) reported that some alleles only appear in Simmental cattle i.e. locus ETH225 allele 158, SPS115 allele 250, TGLA122 alleles 181, and SPS113 allele 289. While for the Simmental population in our study, only locus SPS115 allele 250 and TGLA122 alleles 181 were observed. It is worth noting that these two alleles were observed only in the Simmental Cross cattle population.
Compared to the FAO (2011) recommendation and the results from other studies that also used microsatellites which were mostly identical with our study, some differences can be observed. The differences may be in the minimum and maximum size of allele, the number of observed alleles and also the PIC value. The comparison is presented in Table 3. Differences in the size range of allele and the number of observed alleles in this study were reasonable, due to the characteristics of the microsatellites that are naturally highly polymorphic. The results from Janík et al. (2001) and Czerneková et al. (2006) also confirm that a number of microsatellite markers in the Simmental cattle appeared to be highly polymorphic. Typical occurrence and observation of the private alleles reveal differences that may be an indication for the unique characteristic of the sample used in this study.
The PIC values in this study and some other studies have a high value (PIC>0.5), while the number of observed alleles in this study for the entire microsatellite locus is higher than what is reported by other studies (Table 3). The 12 microsatellite loci with a high PIC value have been reported in other breeds of cattle (Dadi et al., 2008; Kesvulu et al., 2009; Movahedin et al., 2010; Suh et al., 2014). Studies of the Simmental Cross cattle have also been carried out by Czerneková et al. (2006) and revealed a high PIC value and observed heterozygosity values. This enabled the 11 microsatellite loci polymorphism to separate the local breed cattle in the Czech Republic based on genetic distance.
Result of genetic distance analysis shows that the Simmental Cross-Pariaman and the Simmental Cross-Solok subpopulation have the lowest genetic distance value thus it can be interpreted that these subpopulations have a very close genetic relationship. In contrast, the Ongole Grade (PO)-West Java and the Simmental Cross-Pariaman subpopulation have the highest genetic distance value (Table 4). All identified alleles in this study were able to classify the cattle population into groups. There are subpopulations that are closely related and form their own group.
According to the genetic relationship, the Simmental Cross cattle from West Sumatra are divided into two groups. One group is very close to the Simmental Purebred and the other clearly distinct from the Simmental Purebred. Several Simmental Cross subpopulations, including the Pariaman subpopulation (very low population of Simmental cattle) clustering together in one group and separated from the Simmental Purebred population (Figure 2). Meanwhile, the Simmental Cross-West Java subpopulation is distant to the Simmental Cross-West Sumatra and the Simmental Purebred population as well as to the Ongole Grade (PO)-West Java population.
An overview of the genetic relationship on the dendrogram (Figure 2) also reveal that the Simmental Purebred and the Simmental Cross (Agam and Limapuluh Kota subpopulation) are indeed highly related groups. Thus, the Simmental Cross in West Sumatra, especially Agam and Limapuluh Kota subpopulation is the subpopulation that has the closest relationship to the Simmental Purebred cattle. Information about the genetic distance in this study confirms Agung et al. (2014) who reported that the Simmental Cross cattle in Agam and Limapuluh Kota district have a close Mahalanobis distance to the Simmental Purebred cattle based on morphological parameters.
Using STRUCTURE HARVESTER, K optimal was obtained at K = 2. At K = 2, the Simmental Cross-West Sumatra population showed an admixture pattern between the Simmental breed and the Ongole Grade (PO) breed. Furthermore, the Ongole Grade (PO) cattle breed was identified as a separate cluster. The STRUCTURE analysis generate similar interpretation with the dendrogram in Figure 2. The results confirms the hybrid nature of the Simmental Cross-West Sumatra population. In addition, distinct genetic difference in the degree of admixture was observed between the Simmental Cross-West Java and the Ongole Grade (PO) populations. The results from the STRUCTURE analysis also revealed that the Simmental Purebred cattle in our study may not be considered as pure Simmental breed. This may be due to limitation in the number of Simmental purebred individuals studied. The structure analysis is presented in Figure 3.
PCA was used to cluster individual microsatellite genotypes. PCA clusters individuals only on the basis of their genotypes and makes no assumptions regarding the Hardy-Weinberg equilibrium or linkage equilibrium (Rutledge et al., 2010). PCA analysis also indicated a close relationship between the Simmental Cross population and the Simmental Purebred population. Based on the results of genetic distance that represented by the dendrogram in Figure 2 and the PCA analysis results in Figure 4, there was scientific evidence that the Simmental Cross cattle in West Sumatra are the result of grading-up with the Simmental Purebred. In other countries, grading-up the local breed cattle has also been done, for example in the Czech Republic (e.g. Czech Red) which used Simmental cattle to produce the Simmental Cross cattle that genetically had a large distance from their local breed cattle (Czerneková et al., 2006).
The DAPC analysis with assumed K = 4 also indicated the same result with STRUCTURE result (K = 4). However, DAPC assumed a close relationship between the Simmental Cross-West Java and the Simmental breed. This could also indicate that the Simmental Cross-West Java is the result of grading-up with the Simmental Purebred. Moreover, the Ongole Grade (PO) and the Simmental Cross-West Sumatra were identified as separate clusters (Figures 5 and 6).
The value of the genetic distance can be influenced by many factors, including the number of the population used in the study and the objectives of the breeding (Radko et al., 2005) as well as the massive introgression of the breed cattle that was possible because of the geographical relationship (Zhou et al., 2005). This geographical relationship might be one of the influential factors that caused a tight genetic relationship between the Simmental Cross (Agam and Limapuluh Kota subpopulations) in West Sumatra and the Simmental Purebred population. According to the evaluation of the results regarding genetic diversity and advantages possessed by the Simmental Cross cattle, the Simmental Cross population in Agam and Limapuluh Kota can be considered as the initial (base) population for the Simmental Cross cattle breeding programs in West Sumatra, Indonesia.
Information about comparison between the groups of Simmental Cross cattle population in West Sumatra based and the Simmental Purebred cattle based on their genetic distance values provide some benefits for farmers and the local government to design future conservation and breeding programs as well as enrich the local cattle genetic resources.
ACKNOWLEDGMENTS
This study is one of the MeatMilk-Project that funded by the Research Center for Biotechnology-Indonesian Institute of Sciences. We would like to thank the Head of West Sumatra’s Livestock Department, Head of Tuah Sakato Artificial Insemination Center (BIBD Tuah Sakato), Head of Lembang Artificial Insemination Center (BIB Lembang), and Karya Anugerah Rumpin Farm (PT. KAR) for supporting this study
Figure 2
Dendrogram of Simmental Cross-West Sumatra subpopulation using Unweighted Pair-Group Method with Arithmetic mean method based on Nei et al. (1978).
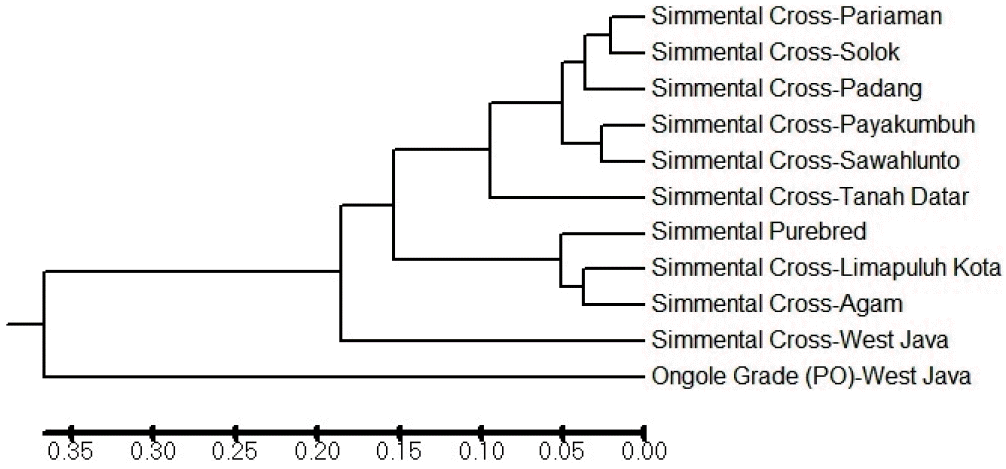
Figure 3
Genetic structures of the Simmental Purebred cattle population (1), the Simmental Cross-West Sumatra (2), the Simmental Cross-West Java (3), and the Ongole Grade (PO) breed (4). Black lines separate individual populations whose names are indicate.
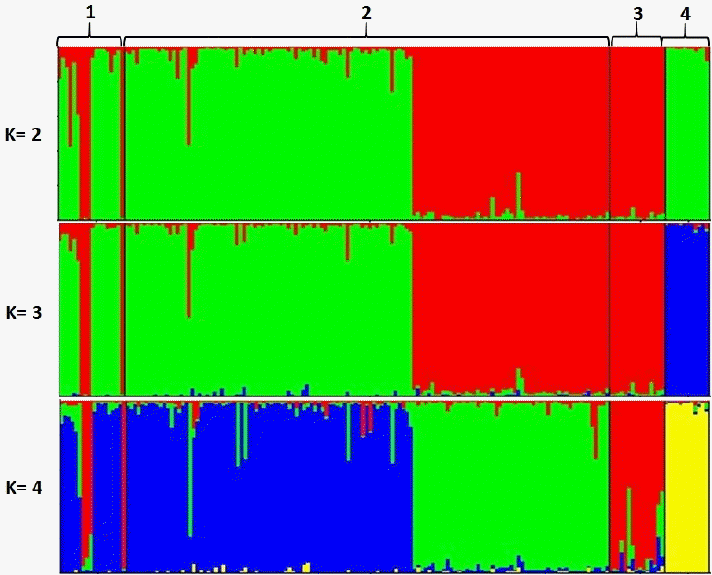
Figure 4
First and third components of a principle components analysis of 12-locus microsatellite genotypes from the Simmental Cross, the Simmental Purebred, and the Ongole population.
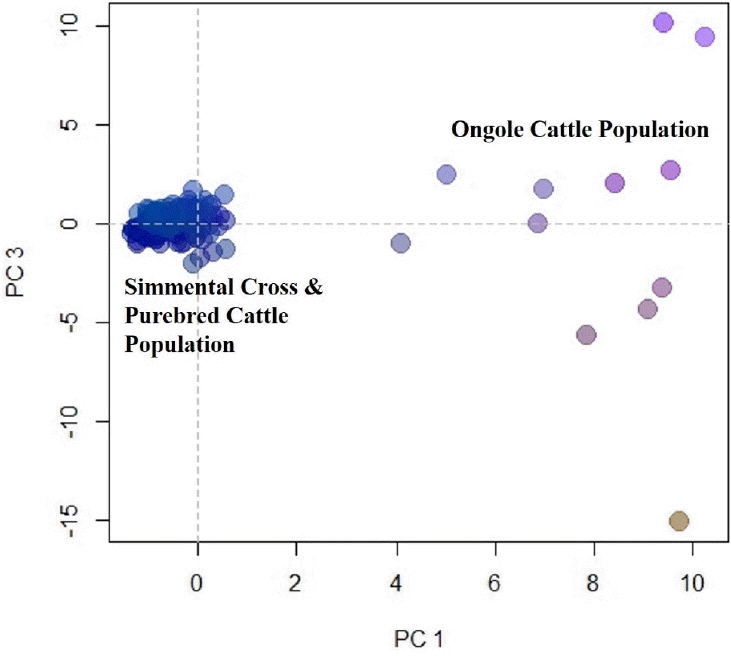
Figure 5
The scatterplot of discriminant analysis of principal components result using 12 microsatellite data. 1, Simmental Purebred; 2, Simmental Cross West Sumatra; 3, Simmental Cross West Java; 4, Ongole Grade (PO) breed. PCA, principal component analysis; DA, discriminant analysis.
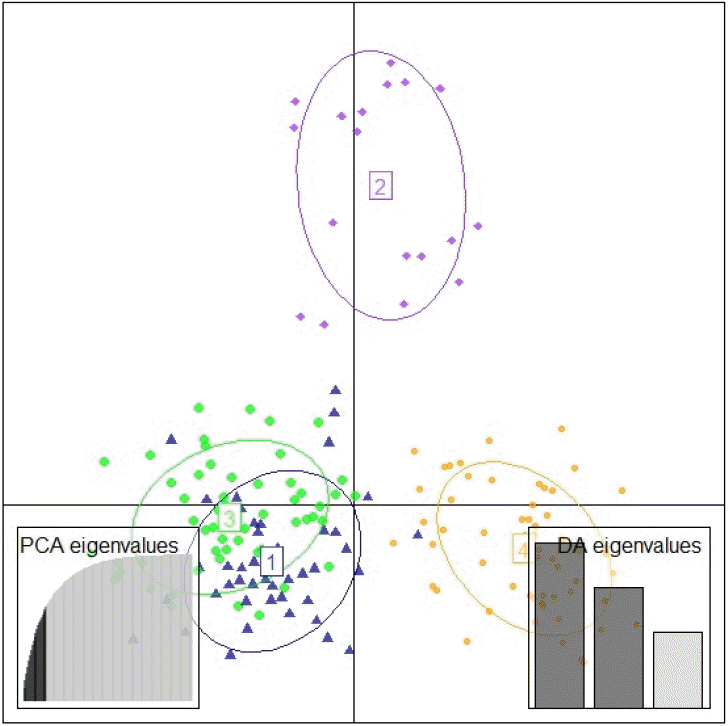
Figure 6
Bayesian approach results using the discriminant analysis of principal components to investigate genetic structure. M18, Simmental Purebred; S131, Simmental Cross-West Sumatra; S146, Simmental Cross-West Java; PO12, Ongole Grade (PO) breed.
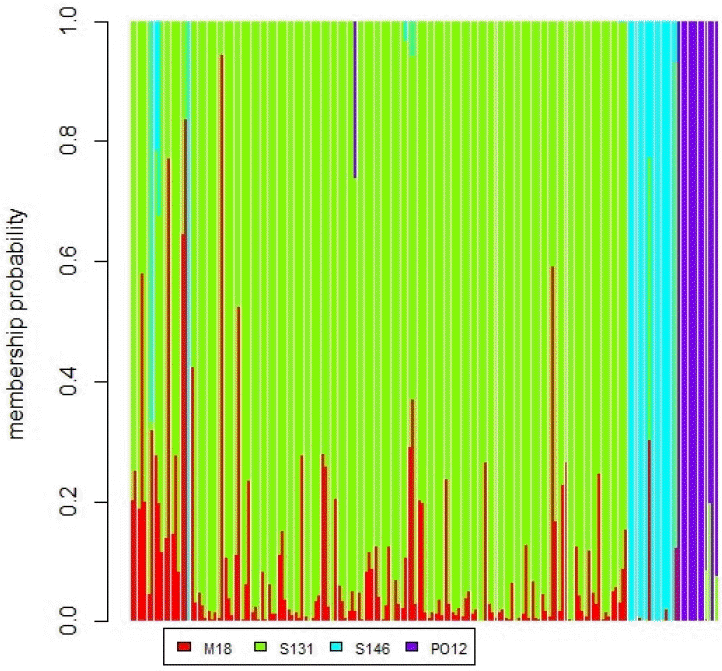
Table 1
Sequence of 12 microsatellite primers used in the study based on FAO (2011)
| Locus | Sequence (5′-3′) | Label | Annealing temp. (°C) | Size of PCR product (base pair) |
|---|---|---|---|---|
| TGLA227 |
F: CGAATTCCAAATCTGTTAATTTGCT R: ACAGACAGAAACTCAATGAAAAGCA |
Fam | 55 | 75–105 |
| SPS113 |
F: CCTCCACACAGGCTTCTCTGACTT R: CCTAACTTGCTTGAGTTATTGCCC |
Hex | 55 | 132–170* |
| BM1824 |
F: GAGCAAGGTGTTTTTCCAATC R: CATTCTCCAACTGCTTCCTTG |
Tamra | 57 | 176–197 |
| ETH225 |
F: ATCACCTTGCCAATATTTCC R: ACATGACAGCCAGCTGCTACT |
Hex | 55 | 131–159 |
| INRA023 |
F: GAGTAGAGCTACAAGATAAACTTC R: TAACTACAGGGTGTTAGATGAACTCA |
Tamra | 55 | 195–225 |
| TGLA122 |
F: CCCTCCTCCAGGTAAATCAGC R: AATCACATGGCAAATAAGTACATAC |
Fam | 57 | 136–184 |
| CSSM66 |
F: ACACAAATCCTTTCTGCCAGCTGA R: AATTTAATGCACTGAGGAGCTTGG |
Hex | 59 | 171–209 |
| ILSTS006 |
F: TGTCTGTATTTCTGCTGTGG R: ACACGGAAGCGATCTAAACG |
Tamra | 59 | 277–309 |
| BM1818 |
F: AGCTGGGAATATAACCAAAGG R: AGTGCTTTCAAGGTCCATGC |
Tamra | 59 | 248–278 |
| SPS115 |
F: AAAGTGACACAACAGCTTCTCCAG R: AACGAGTGTCCTAGTTTGGCTGTG |
Fam | 57 | 234–258 |
| TGLA126 |
F: CTAATTTAGAATGAGAGAGGCTTCT R:TTGGTCTCTATTCTCTGAATATTCC |
Fam | 58 | 115–131 |
| TGLA53 |
F: CAGCAGACAGCTGCAAGAGTTAGC R: CTTTCAGAAATAGTTTGCATTCATGCAG |
Hex | 51 | 143–191 |
Table 2
Summary statistic of the mean number of observed allele (Na), mean number of effective alleles (Ne), observed (Ho) and expected (He) heterozygosities observed in 11 cattle populations
Table 3
Comparison of size range of microsatellite alleles in Simmental cattle
| Locus | Parameter | Choroszy et al. (2006) | Jevrosima et al. (2009) | Riojas-Valdes et al. (2009) | This study |
|---|---|---|---|---|---|
| TGLA227 | Range (bp) [nA] | 77–97 [10] | 78–114 [11] | 76–98 [13] | 71–99 [12] |
| Ho | 0.89 | 0.733 | 0.9301 | 0.724 | |
| PIC | 0.824 | 0.82 | 0.9999 | 0.842 | |
| BM1824 | Range (bp) [nA] | 178–188 [4] | 176–190 [6] | – | 177–195 [7] |
| Ho | 0.725 | 0.71 | - | 0.591 | |
| PIC | 0.607 | 0.7 | - | 0.694 | |
| ETH225 | Range (bp) [nA] | 140–152 [6] | 134–150 [7] | 142–162 [5] | 134–164 [13] |
| Ho | 0.769 | 0.594 | 0.7058 | 0.767 | |
| PIC | 0.642 | 0.68 | 0.9809 | 0.801 | |
| INRA23 | Range (bp) [nA] | 198–218 [8] | 198–220 [11] | - | 192–218 [12] |
| Ho | 0.659 | 0.774 | - | 0.696 | |
| PIC | 0.665 | 0.86 | - | 0.747 | |
| TGLA122 | Range (bp) [nA] | 143–161 [5] | 136–162 [9] | - | 135–185 [17] |
| Ho | 0.703 | 0.452 | - | 0.559 | |
| PIC | 0.591 | 0.7 | - | 0.678 | |
| SPS115 | Range (bp) [nA] | 248–262 [8] | 242–254 [6] | 250–265 [11] | 242–264 [8] |
| Ho | 0.667 | 0.719 | 0.7983 | 0.578 | |
| PIC | 0.615 | 0.6 | 0.9934 | 0.627 | |
| TGLA126 | Range (bp) [nA] | 113–123 [5] | 114–124 [6] | - | 104–130 [10] |
| Ho | 0.67 | 0.667 | - | 0.604 | |
| PIC | 0.596 | 0.59 | - | 0.658 | |
| TGLA53 | Range (bp) [nA] | 146–190 [16] | 150–184 [14] | - | 128–168 [19] |
| Ho | 0.747 | 0.656 | - | 0.742 | |
| PIC | 0.851 | 0.88 | - | 0.877 |
Table 4
Genetic distance value in 11 cattle population (unbiased measures of genetic identity and genetic distance based on Nei et al. [1978])
A, Simmental Purebred; B, Simmental Cross-Limapuluh Kota; C, Simmental Cross-Agam; D, Simmental Cross-Payakumbuh; E, Simmental Cross-Pariaman; F, Simmental Cross-Padang; G, Simmental Cross-Tanah Datar; H, Simmental Cross-Sawahlunto; I, Simmental Cross-Solok; J, Simmental Cross-West Java; K, Ongole Grade (PO)-West Java.
REFERENCES
Agung PP, Ridwan M, Indriawati , Handrie , Saputra F, Supraptono , Erinaldi . 2014. Morphological profile and estimation of genetic distance of simmental crossbred. Indonesian J Anim Vet Sci 19:112–122.
Cervini M, Henrique-Silva F, Mortari N, Matheucci E. 2006. Genetic variability of 10 microsatellite markers in the characterization of Brazilian Nellore cattle (Bos indicus). Genet Mol Biol 29:486–490.

Choroszy B, Janik A, Choroszy Z, Ząbek T. 2006. Polymorphism of selected microsatellite DNA sequences in Simmental cattle chosen for identification of QTLs for meat traits. Anim Sci Pap Rep 24:Suppl 271–77.
Czerneková V, Kott T, Dudková G, Sztankóová Z, Soldát J. 2006. Genetic diversity between seven central European cattle breeds as revealed by microsatellite analysis. Czech J Anim Sci 51:1–7.

Dadi H, Tibbo M, Takahashi Y, Nomura K, Hanada H, Amano T. 2008. Microsatellite analysis reveals high genetic diversity but low genetic structure in Ethiopian indigenous cattle populations. Anim Genet 39:425–431.


Earl DA, vonHoldt BM. 2012. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genet Resour 4:359–361.

Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol 14:2611–2620.


FAO. 2011. Molecular genetic characterization of animal genetic resources. FAO Animal Production and Health Guidelines. No. 9. http://www.fao.org/docrep/014/i2413e/i2413e00.htmAccessed February 10, 2015
Glaubitz JC. 2004. Convert: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes 4:309–310.

Janík A, Žábek T, Radko A, Natonek M. 2001. Evaluation of polymorphism at 11 microsatellite loci in Simmental cattle raised in Poland. Ann Anim Sci 1:19–29.
Jevrosima S, Stanimirovic Z, Dimitrijevic V, Stojic V, Fratric N, Lazarevic M. 2009. Microsatellite DNA polymorphism and its usefulness for pedigree verification in Simmental cattle from Serbia. Acta Vet (Beograd) 59:621–631.

Jombart T. 2008. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405.


Kalinowski ST, Taper ML, Marshall TC. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106.


Kesvulu PC, Rao GN, Ahmed ASN, Gupta BR. 2009. Molecular genetic characterization of Punganur cattle. Indian J Vet Anim Sci Res 5:179–185.
Mao Y, Chang H, Yang Z, Zhang L, Xu M, Chang G, Sun W, Song G, Ji D. 2008. The analysis of genetic diversity and differentiation of six Chinese cattle populations using microsatellite markers. J Genet Genomics 35:25–32.


Maretto F, Ramljak J, Sbarra F, Penasa M, Mantovani R, Ivankovic A, Bittante G. 2012. Genetic relationships among Italian and Croatian Podolian cattle breeds assessed by microsatellite markers. Livest Sci 150:256–264.

Movahedin MR, Amirinia C, Noshary A, Mirhadi SA. 2010. Detection of genetic variation in sample of Iranian proofed Holstein cattle by using microsatellite marker. Afr J Biotechnol 9:9042–9045.
Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590.




Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959.




Putnova L, Vrtkova I, Srubarova P, Stehlik L. 2011. Utilization of a 17 microsatellites set for bovine traceability in Czech cattle populations. Iranian J Appl Anim Sci 1:31–37.
R Development Core Team. 2015. R: A language and environment for statistical computing. http://www.R-project.orgAccessed April 28, 2015
Radko A, Zyga A, Zabek T, Slota E. 2005. Genetic variability among Polish Red, Hereford and Holstein-Friesian cattle raised in Poland based on analysis of microsatellite DNA sequences. J Appl Genet 46:89–91.

Rehman MS, Khan MS. 2009. Genetic diversity of Hariana and Hissar cattle from Pakistan using microsatellite analysis. Pakistan Vet J 29:67–71.
Riojas-Valdes VM, Gomes-de-la-Fuente JC, Garza-Lozano JM, Gallardo-Blanco DC, De Tellitu-Schutz JN, Wong-Gonzales A, Davalos-Aranda G, Salinas-Melendez JA. 2009. Exclusion probabilities of 8 DNA microsatellites in 6 cattle breeds from Northeast Mexico. J Anim Vet Adv 8:62–66.
Rutledge LY, Garroway CJ, Loveless KM, Patterson BR. 2010. Genetic differentiation of eastern wolves in Algonquin Park despite bridging gene flow between coyotes and grey wolves. Heredity 105:520–531.


Siregar AR, Bestari J, Matondang RH, Sani Y, Panjaitan H. 1999. Penentuan sistem breeding sapi potong program IB di propinsi Sumatera Barat. http://peternakan.litbang.pertanian.go.id/english/index.php?option=com_content&view=article&id=3063%3Asemnas&catid=310%3Asemnas1999&Itemid=121Accessed January 15, 2015
Statistics Indonesia. 2011. Data collection beef cattle, dairy cattle, and buffalo 2011 (PSPK2011). http://www.bps.go.id/index.php/publikasi/681
Suh S, Kim Y-S, Cho C-Y, Byun M-J, Choi S-B, Ko Y-G, Lee CW, Jung K-S, Bae KH, Kim J-H. 2014. Assessment of genetic diversity, relationships and structure among Korean native cattle breeds using microsatellite markers. Asian Australas J Anim Sci 27:1548–1553.



Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729.



Yeh FC, Boyle TJB. 1997. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129:157–163.





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





