 |
 |
Abstract
This study investigated the geographic and pairwise distances of nine Chinese local Cashmere goat populations through the analysis of 20 microsatellite DNA markers. Fluorescence PCR was used to identify the markers, which were selected based on their significance as identified by the Food and Agriculture Organization of the United Nations (FAO) and the International Society for Animal Genetics (ISAG). In total, 206 alleles were detected; the average allele number was 10.30; the polymorphism information content of loci ranged from 0.5213 to 0.7582; the number of effective alleles ranged from 4.0484 to 4.6178; the observed heterozygosity was from 0.5023 to 0.5602 for the practical sample; the expected heterozygosity ranged from 0.5783 to 0.6464; and Allelic richness ranged from 4.7551 to 8.0693. These results indicated that Chinese Cashmere goat populations exhibited rich genetic diversity. Further, the Wright’s F-statistics of subpopulation within total (FST) was 0.1184; the genetic differentiation coefficient (GST) was 0.0940; and the average gene flow (Nm) was 2.0415. All pairwise FST values among the populations were highly significant (p<0.01 or p<0.001), suggesting that the populations studied should all be considered to be separate breeds. Finally, the clustering analysis divided the Chinese Cashmere goat populations into at least four clusters, with the Hexi and Yashan goat populations alone in one cluster. These results have provided useful, practical, and important information for the future of Chinese Cashmere goat breeding.
Goats occupy an extremely important position in human history as one of the first animal species to be domesticated (Luikart et al., 2001; Fernandez et al., 2006). Goats possess a special ability to adapt rapidly to different environmental conditions, and the species’ special characteristics put it in a position of economic importance. Cashmere goats are found only in specific areas of Asia, with modern Cashmere goat breeds generally originating from the territorial plateau of southwestern China and adjacent areas of central Asia (Tu, 1989). The world’s largest number of Cashmere goat populations is raised in China, and these populations are a significant resource: more than ten different populations have developed after long-term natural and human selection (Qi, 2004). Among these populations, the Liaoning and Inner Mongolian Cashmere goats (including three types of Alashan, Erlangshan, and Wuzhumuqin) are well known worldwide for their high cashmere yield and cashmere fineness, respectively. Cashmere goat breeding plays an important role in Chinese animal agriculture, and particularly so in underdeveloped, rural, and mountain areas, where production systems are based on local or native breeds. In the past three decades, a high cashmere yield has been the main focus in breeding. Importantly, genetic admixture among some breeds, as well as inbreeding within the breed/types, is possible. These activities may have led to a loss of genetic structure and diversity in some of these breed/types. In recent decades, an increasing focus on cashmere production within animal agriculture has resulted in changes in Cashmere goat management systems: there is an increasing emphasis on the breeding of a small number of very productive cosmopolitan breeds. In light of this, it is of great importance to conserve genetic resources in order to maintain more extensive breeding options. The effective assessment of genetic variability in domestic Cashmere goat populations constitutes the first step toward this genetic resource conservation.
Phylogenetic studies of population diversification have allowed researchers to identify the essential features in many species’ evolutionary histories (Avise, 2000). Population differentiation through reduced gene flow, selection, and genetic drift can alter the geographic species shift scenarios that occur under regional warming (Davis and Shaw, 2001). The intensification of production systems and the successes observed in industrial breeding have led some farmers to abandon certain native breeds. Additionally, domesticated animals are presently experiencing a loss of genetic diversity as a result of several other factors. First, the intensive selection of highly productive breeds has placed inadequate emphasis on the preservation of overall genetic diversity. Second, native breeds in marginal areas are facing extinction, and little or no action is being taken to reverse this trend (Taberlet et al., 2008). Microsatellites provide accurate genetic information on individual genotypes and the genetic relationships between populations (Arranz et al., 1998; Ruane, 1999; Kantanen et al., 2000). However, for analysis of microsatellite data, the efficiencies of distance measures in phylogenetic reconstruction as compared to the traditional distance metrics remain unknown. In this paper, we will consider only one neighbor-joining (NJ) method (Saitou and Nei, 1987) that is assessed using genetic distances (DS) and (DA), because this particular NJ method is considered to be robust across many situations (Nei, 1991). The calculation of genetic distances using microsatellite data can provide useful information for the overall monitoring and management of the genetic diversity of rare breeds.
Microsatellite DNA is currently the most useful and widely used marker that can be used to perform a wide range of molecular genetic studies, including establishing the population structure (Bruford, 1993), population differentiation, and reconstruction of phylogenetic relationships among populations (Bowcock, 1994; MacHugh, 1997; Wimmers, 2000). These microsatellites provide accurate genetic information on individual genotypes and the genetic relationships between populations (Arranz et al., 1998; Ruane, 1999; Kantanen et al., 2000). Such genetic diversity analyses can provide insight into domestication events, relationships among breeds, loss of within-breed genetic diversity, breed structure, etc., and are essential for establishing conservation priorities (Toro et al., 2009). In recent years, efforts have been made to carry out studies that are aimed to characterize the genetic diversity in goat breeds from several different countries and regions; the available information has been reviewed recently (Groeneveld et al., 2010). The vast majority of these genetic diversity studies has used microsatellite markers to obtain information, and most of the studies cover a sample of breeds from a given country (Ganai and Yadav, 2001; Li et al., 2002; Tadlaoui-Ouafi et al., 2002; Visser et al., 2004; Xiang-Long and Valentini, 2004; Iamartino et al., 2005; Sechi et al., 2005; Araújo et al., 2006; Gour et al., 2006; Martínez et al., 2006; Oliveira et al., 2007; Fatima et al., 2008; Glowatzki-Mullis et al., 2008; Wang et al., 2008; Qi et al., 2009; Traoré et al., 2009; Bruno-de-Sousa et al., 2011; Ozgecan and Okan, 2012). Some studies, however, have considered a broader geographical scope, including larger samples of breeds from Asia (Barker et al., 2001), the Mediterranean region (Canon et al., 2006), Africa (Muema et al., 2009), and breeds from multiple continents (Martinez-Martinez et al., 2008; Oliveira et al., 2010; Di et al., 2011; Gamaa and Bressan, 2011). Furthermore, Di et al. revealed that Chinese Cashmere goat breeds/types exhibited a rich genetic diversity (2011), with expected heterozgosity and allelic richness values ranging from 0.54 to 0.65 and from 3.90 to 5.96 respectively. Further, Di et al. showed that the expected heterozgosity of Chinese Cashmere goats was similar to that of West African goats, but lower than that of Iranian goats. The aim of the present study was to assess how useful were microsatellite polymorphisms for analysis of the genetic relationships between nine Chinese local Cashmere goat populations, with the ultimate aim of maintaining and conserving the populations. Results of this analysis will help to further understand the morphology and geographical distribution among Chinese Cashmere goat populations.
We selected nine Chinese local Cashmere goat populations for investigation in this study. For analysis, 10 ml blood samples were collected from the jugular vein of each investigated animal. From the 10 ml samples, 2 ml samples were quickly frozen in liquid nitrogen and stored at −80°C for genomic DNA extraction, as described previously (d’Angelo et al., 2006). The total DNA was extracted from whole blood using the saturated salt method (Sambrook et al., 1989), quantified spectrophotometrically, and adjusted to 50 ng/μl. Blood samples were collected from 809 goats from nine breeds/types, spanning the entire distribution range of Chinese Cashmere goats. The genetic characteristics for these Cashmere goats were analyzed in order to ascertain the historical relationships and relative genetic contributions for the populations. The investigated populations included the following numbers and corresponding population types: 84 Hexi Cashmere goat (HX), 108 Chaidamu Cashmere goat (CDM), 112 Xinjiang Cashmere goat (XJ), 116 Liaoning Cashmere goat (LN), 96 Erlangshan Cashmere goat (ELS), 72 Alashan Cashmere goat (ALS), 76 Hanshan Cashmere goat (HS), 86 Wuzhumuqin Cashmere goat (WZMQ), and 59 Yashan Cashmere goat (YS) raised in China (Table 1).
Primers flanking 20 microsatellite loci located at several different chromosomes were designed by an available genome sequence (Table 2) (Crawford et al., 1995) and synthesized by the Shanhai Shenggong Biological Engineering Company. The 20 selected microsatellite DNA markers were recommended by the Food and Agriculture Organization of the United Nations (FAO) and the International Society for Animal Genetics (ISAG). PCR reactions took place in a 25 μl reaction system containing 2 μl genomic DNA template; 1.5 μl MgCl2 (25 mmol/L); 2 μl dNTP (10 mmol/L); 0.25 μl Taq DNA polymerase (5 μl/U) (TaKaRa, China); 2 μl each forward and reverse primers (8 pmol/μl); 2.5 μl 10×PCR Buffer; and ddH2O (up to 25 μl). The PCR conditions were as follows: initial denaturation for 5 min at 94°C, 35 cycles at 94°C for 1 min, annealing temperatures at 52 to 58°C for 40 s, and extension at 72°C for 40 s. The final extension step was followed by a 5 min extension at 72°C.
For analysis, 2 μl PCR product was mixed with 6 μl denaturing solution (95% formamide, 25 mM EDTA, 0.025% xylenecyanole, and 0.025% bromophenol blue), heated for 10 min at 98°C, and then chilled on ice. The samples were electrophoresed with 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The electrophoresis ran at 250 V and 40 mA (pre-electrophoresis) for 10 min, followed by 150 V and 24 mA (Kucharczyk Techniki Elektroforetyczne) for 8 h (silver staining). A refrigerated circulator was used to control the gel temperature (4°C). Then, each locus of the number of alleles was computed using Excel Microsatellite Toolkit (Version 3.1) software. The fragment length of PCR products was determined using GeneMapper software (Version 3.7).
The gene and genotypic frequencies were estimated using direct gene counting. The Na, Ne (Kimura and Crow, 1974), Pa, He (Nei et al., 1983), Ho, PIC (Botstein et al., 1980), and AR values were estimated using POPGENE software (version 1.31) (Yeh et al., 1997). The significance of the F-statistics was determined from permutation tests, in which the sequential Bonferroni procedure was applied over loci (Hochberg, 1988). The F-statistics indices (Wright, 1978) were computed using the FSTAT program (Goudet, 2002), and the estimate was based on the following relationship: Nm = 0.25(1−FST)/FST, where FST was the mean FST value across all loci (Slatkin and Barton, 1989). Reynolds’ genetic distance between populations was subsequently calculated on the basis of the FST values (Reynolds et al., 1983). A consensus neighbour-joining (NJ) dendrogram of the nine Chinese local Cashmere goat populations, based on Nei′s genetic distances (DS) (Nei, 1972) and Nei′s genetic distances (DA) (Nei et al., 1983), was reconstructed using DISPAN software (version 1.1); this was the most effective way to obtain accurate phylogenetic trees (Saitou and Nei, 1987; Takezaki and Nei, 1996). Finally, GST was calculated on the basis of the data from all loci (Takezaki et al., 2010).
Table 2 and 3 display the following data concerning the 20 microsatellite loci used from the nine Chinese local Cashmere goat populations: a description of the markers, including the chromosomal localization, the number of alleles per marker, and the fragment sizes; observed and expected heterozygosities; the heterozygote deficiency within population (FIS); and the deviation from the Hard-Weinberg equilibrium. The 20 microsatellites included a total of 206 alleles, and the mean number of alleles per locus in the population was 10.30. Most of the markers used had allele number values that were higher than nine, and the values ranged from seven (SRCRSP3, OarFCB48, BMS1494, and BM1818) to 16 (DRBP1) alleles per marker. The fragment sizes ranged from 107 bp to 123 bp, from 138 bp to 178 bp, from 227 to 237 bp, from 200 to 284 bp, and from 106 to 154 bp, respectively (Table 2).
Table 3 contains genetic variation metrics for the nine Chinese local Cashmere populations. The highest mean numbers of effective alleles were found in LN (8.4025) and XJ (8.3608), followed by CDM (7.9613) and ELS (7.8812). Meanwhile, YS had the lowest mean number of effective alleles (6.7683). The highest average numbers of effective alleles were found in XJ (4.6178) and LN (4.5649), followed by ELS (4.4936) and CDM (4.3350); meanwhile, the lowest average number of effective alleles was found in YS (4.0484). The genetic diversity difference index (PIC value) for the whole dataset was high (0.6500), and most of the populations had PIC values above 0.6198. The lower PIC values were present in YS (0.5213) and WZMQ (0.5468), while higher PIC values were observed for XJ and LN (0.7582 and 0.7187, respectively). The observed heterozygosity (Ho) averaged over all loci was 0.5249, while the expected heterozygosity (He) was 0.6084. The allelic richness in the nine populations ranged from 4.7551 to 8.0693; it was higher in XJ and LN populations than in YS Cashmere goat populations (Table 3). Therefore, the 20 microsatellite loci could be used to evaluate the genetic diversity in Cashmere goat populations. Furthermore, results demonstrated that the investigated Chinese Cashmere goat populations exhibited a rich genetic diversity.
The genetic structure and genetic variation of the Cashmere goat populations were analyzed using the 20 microsatellite loci along with Wright’s F-statistics. The mean FIT was 0.1374, the mean FST was 0.1184, and the mean FIS was 0.0229. Meanwhile, the GST values ranged from 0.0273 to 0.1738; Nm values for the markers varied from 1.0171 to 3.7246; the mean GST was 0.0940; and the mean Nm was 2.0415. The variation results both among and within populations for the small number of individuals can be characterized as inbred. Meanwhile, the distribution of the nine Chinese Cashmere goat populations differed in various microsatellite loci (p<0.05, p<0.01 or p<0.001) (Table 4).
The genetic phylogeny was analyzed using three methods described by Peter et al. (2007). We determined an optimum K value of 4, following Evanno et al. (2005). Evanno et al. found that an ad hoc quantity, which was based on the second order rate of change of the likelihood function with respect to K, did show a clear peak at the true value of K. Meanwhile, Pritchard and Falush have developed methods that allow for linkage between loci, which can produce highly accurate assignments using modest numbers of loci, (Pritchard et al., 2000; Falush et al., 2003). A clear clustering (K = 4) was observed, which was in accordance with population types (Figure 1). Further, the genetic distance between populations was analyzed using Nei′s genetic distance (DA) and Nei′s standard genetic distance (DS), and neighbour-joining phylogenetic trees for both methods were similar (Figures 2 and 3). More specifically, the neighbour-joining dendrogram that was based on Nei′s genetic distances effectively divided the nine Chinese local Cashmere goat populations into four groups. YS, which produces very little cashmere, was genetically distinct and was the first to separate. The other Chinese goats were then divided into three sub-clusters. The first cluster included LN, CDM, and XJ, which belonged to the high-yield-cashmere type. The second cluster included ALS, ELS, WZMQ, and HS, which belonged to the fine-cashmere type. The third cluster included HX, and was farther from all of the other Chinese goats, with the exception of YS. The three-factor correspondence analysis for frequency distributions of the 20 microsatellite alleles in the nine Chinese local Cashmere goat populations is shown in Figure 4. The first two factors accounted for 31.42% and 25.52% of the total variation, respectively, and clearly distinguished the following principal components: Axis I (ELS, ALS, WZMQ, YS, and HS), Axis II (XJ, CDM, and LN), and Axis III (HX). The first two factors fit well with the geography, while the third factor, contributing 11.77% of the total variation, played an important role in discriminating the Hexi Cashmere goat population (Figure 4).
Table 5 presents the genetic distance and pairwise distance among the nine investigated populations, as shown by DA and DS values. The DA values ranged from 0.0673 to 0.2873, and the DS values from 0.0428 to 0.2745. The genetic distance between LN and CDM was the smallest (DA = 0.0673, DS = 0.0428), and the genetic distance between CDM and YS was the largest (DA = 0.2873, DS = 0.2745).
The polymorphism information content (PIC) for a population, and the heterozygosity of the individuals within, can reflect the extent of genetic variation, which shows high value for large. (Botstein et al., 1980). Information on the genetic variation within the populations investigated in this study is reported in Table 3. This study was expected to provide the genetic evidence for the classification of these breeds/types. The results showed that for the nine Chinese Cashmere goat populations investigated here, He and AR values ranged from 0.5783 to 0.6464 and from 4.7551 to 8.0693, respectively. The lowest value was found in YS and the highest in XJ and HX. The He observed for Chinese goats was similar to that which was reported in previous studies on Chinese goats (Li and Alessio, 2004; Chen et al., 2006; Kang et al., 2011) and West African goats, but lower than that observed in Iranian goats. Further, the allelic richness of the Chinese Cashmere goat populations studied here was higher than the allelic richness reported in other studies. And, the present study included more individuals (809) and more microsatellite markers (20) than the number of individuals (376) and microsatellite markers (14) included in the study by Di et al. Although we conducted similar analyses and used some of the same local populations (6 populations are the same, 3 populations are not the same), the allelic diversity and genetic diversity that resulted here was not consistent with that reported in the prior study (Di et al., 2011). In addition, the present study included PIC, FIT, FIS, GST, and Nm, while previous studies included a more limited set of parameters (Di et al., 2011). Regarding the population cluster produced in this study, Cashmere goat populations from Inner Mongolia were classified into one branch, and other Cashmere goats from Northern China were grouped into a second branch. This clustering further illustrated that in China, there were differences between Cashmere goats that were consistent with genetic distance and geographic distance.
The PIC value was originally introduced by Botstein et al. (1980). Based on the number of detectable alleles and the distribution of their frequency, PIC can indicate how useful a marker can be for detecting polymorphism within a population. Additionally, the PIC value has proven to be a general measure of how informative is a marker (Guo and Elston, 1999): the higher the PIC value, the more the informative the marker. In some cases, populations that have similar frequencies at certain microsatellite loci may still have adaptively important differences that have been maintained by natural selection (Hedrick, 1999). In other cases, populations that have different allele frequencies at certain microsatellite loci may share adaptively important traits.
In general, differentiation at microsatellite loci should reflect the potential for adaptive differences among populations (Gutierrez-Espeleta et al., 2000). In this study, estimates of heterozygosity within the populations were based on a set of microsatellites that indicated that XJ had the largest genetic variability, while YS had the lowest. At the same time, the mean number of alleles and the mean observed and expected heterozygosities of XJ and YS were similar (Table 3). This may be because XJ included a large number of individuals and a broad distributing area, while YS existed in a remote area and had a small population size. Thus, there could have been less gene exchange between YS and other populations. YS ultimately showed an allelic diversity and genetic diversity that was lower than corresponding values for the other eight local Chinese Cashmere goat populations.
In general, Inner Mongolian Cashmere goats have been considered to be one breed. However, all of the obtained pairwise FST values were highly significant (p<0.001 or p<0.01) at 20 microsatellite loci. This indicated that, for breeding purposes, the ALS, ELS, and WZ breeds from Inner Mongolia could be considered independent populations. According to the FST values and the structure plot, the Shandong YS was the most distinct of the Chinese Cashmere goats. Notably, a marked deviation of the Hardy-Weinberg proportions was observed for the marker, which may be explained by the following. The observed null alleles that led to a high within-breed global FST value of 0.1184 for Chinese Cashmere goats in this study was similar to that which was observed for Chinese goats (0.0629 to 0.1632) (Li et al., 2004; Di et al., 2011), higher than that observed for European and Middle Eastern goats (0.069) (Canon et al., 2006), and lower than that observed for Southeast Asian goats (0.143) (Barker et al., 2001). The information offered by GST in the present study allowed for a better differentiation among populations, and this was especially true for the Cashmere goat populations. The observed GST for Cashmere goats showed a high within breed genetic variability characterized by the lowest within breed. To determine whether the degree of scatter shown in the scatter plots increased with geographic distance and whether the populations were in drift-gene flow equilibrium, we correlated the residuals from the FST linear regression with geographic distance. Ultimately, these parameters do not allow gene flow patterns among populations to be ascertained. It has been suggested that the typical high within population variability of microsatellites may result in a low magnitude of differentiation measures (Hedrick, 1999; Balloux and Lugon-Moulin, 2002). This provides a useful context for the FIT vales from this study: 0.0627 to 0.2534, as well as those of FIS: 0.088 to 0.1078. In addition, the most commonly used estimators of gene flow, such as Nm = 0.25(1−FST)/FST, (1.0171 to 3.7246 for this study), are derived on the basis of simplified models of population structure that assume constant population sizes, symmetrical migration at constant rates, and population persistence for periods long enough for achievement of genetic equilibrium (Wright, 1969). These shortcomings highlight the need for the application of new, more informative methodologies in an effort to effectively ascertain the evolutionary history of present-day populations, both for long term gene flow and for the recent migration patterns (Wilson and Rannala, 2003). Here, we further emphasize the need to apply more informative methodologies to ascertain the evolutionary history of current populations. In addition, we present methodologies for the estimation of recent migration patterns to furnish complementary information, thus allowing recent introgression processes to be ascertained.
The observed genetic relationships among the investigated populations have been illustrated using the NJ topology tree derived from the Nei (1978) standard genetic distance. Although the NJ topology tree is not well supported by the nodes, the dendrogram (Figures 2 and 3) shows a clear separation of the Chinese local Cashmere goat populations from different geographic locations. Similar results were observed in the phylogenetic trees of neighbour-joining dendrogram analysis using Nei′s genetic distances (DA) and Nei′s standard genetic distances (DS), which indicated that the phylogenetic relationships among the nine Cashmere goat populations were not closely linearly correlated with their geographic distribution. This result was in agreement with the finding of previous studies, which showed that these populations were originally different, but shared the same genetic material because of natural and artificial selection, as well as their ecological habitat (Li et al., 2002; Chen et al., 2006; Kang et al., 2011). From the perspective of conservation genetics, Hedrick and Miller (1992) have indicated that populations should be managed in a way that retains adequate genetic variability, so as to provide for future adaptation and the successful expansion of native and reintroduced free-ranging populations. Because we cannot directly evaluate the biological significance of the genetic differences between locations, and because genetic differences are roughly proportional to geographic distances, the most conservative method of selecting stock for translocation would be to choose the closest available population to preserve local variation and/or potential adaptation (Gutierrez-Espeleta et al., 2000). According to our investigation, the individual exchanges among Cashmere goat breeds with the aim of increasing cashmere quantity (Li et al., 2004; Wang et al., 2008) might have led to a closer relationship among Cashmere goat populations. This study showed significant levels of genetic divergence between the nine Chinese local Cashmere goat populations, and these findings indicate that the genetic process was associated with historical ecogeographic barriers. To maintain the present genetic diversity and structure of these populations, genetic exchanges between populations must be carefully controlled on existing conservation farms.
ACKNOWLEDGMENTS
This work was supported by the Earmarked Fund for Modern China Wool & Cashmere Technology Research System (No.nycytx-40-2), National Key Technology R & D Program (2011BAD28B05-1), the Gansu Key Technology R & D Program (201104NKCA083), and the Central Level, Scientific Research Institutes for Basic R & D Special Fund Business (1610322012006).
Figure 1.
The model choice criterion ln P(D) (±SD) of the Structure analysis for K values ranging from 2 to 20 for the admixture model.
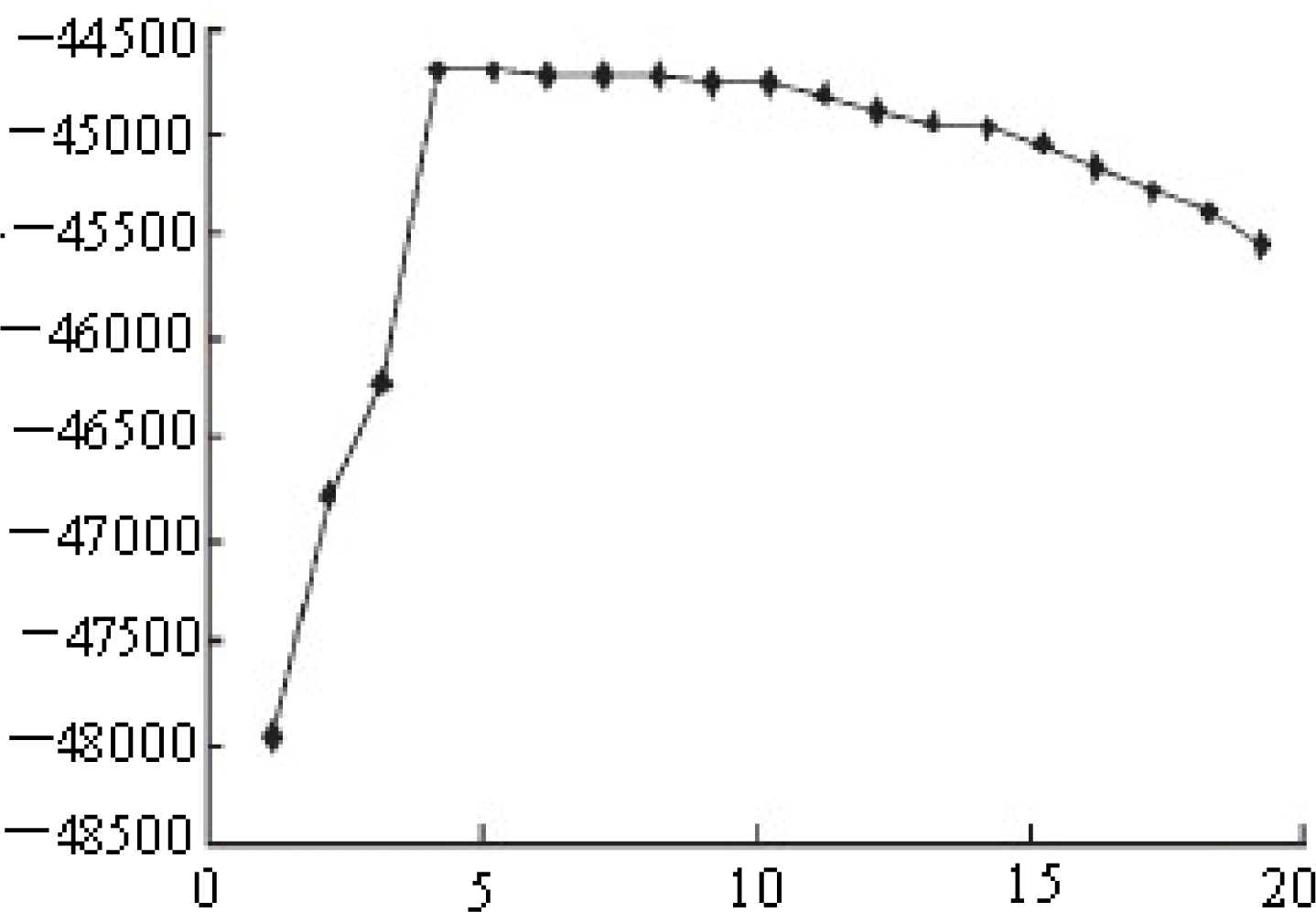
Figure 2.
A neighbour-joining dendrogram of nine Cashmere goat populations based on Nei′s genetic distances (DA).
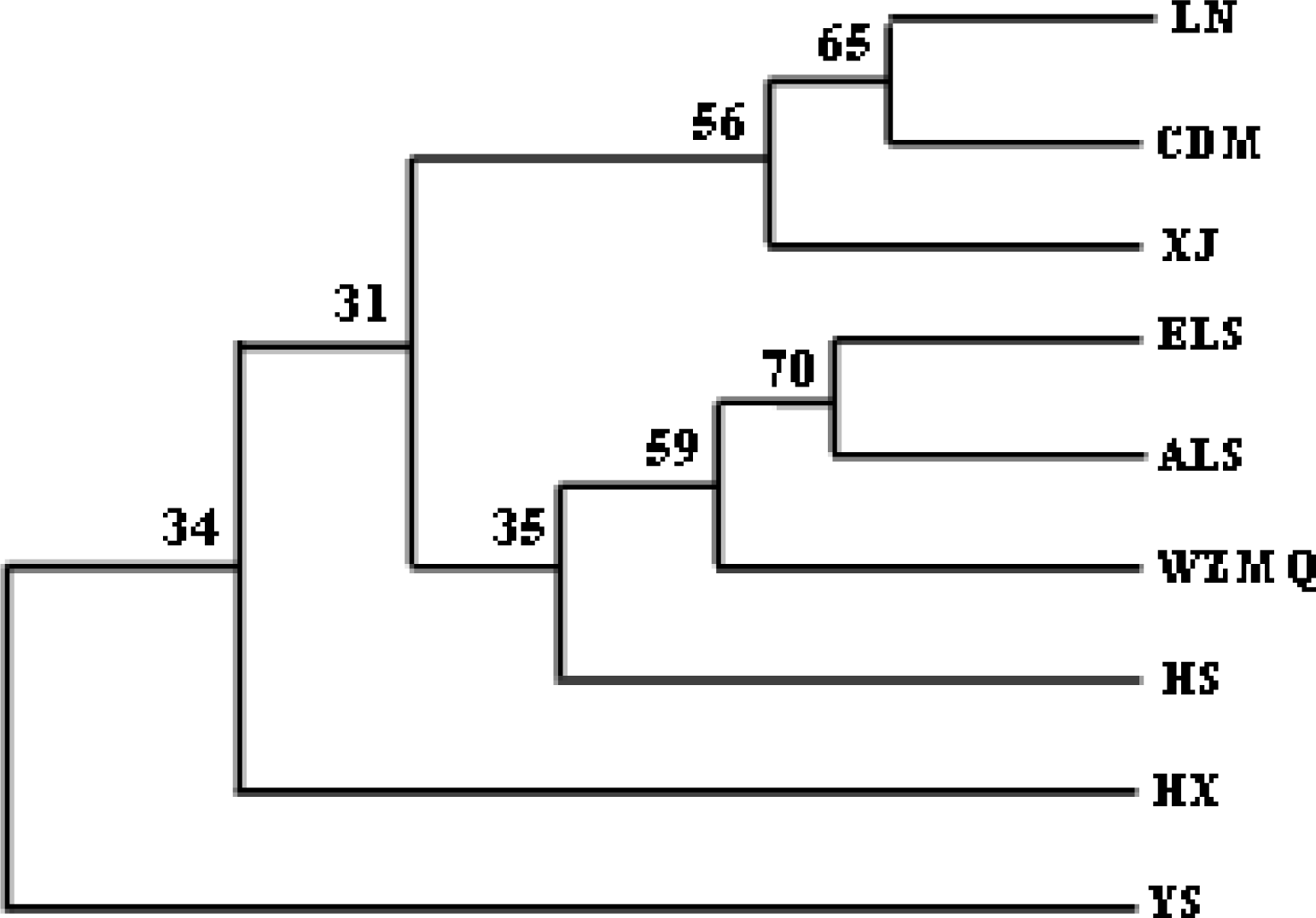
Figure 3.
A neighbour-joining dendrogram of nine Cashmere goat populations based on Nei′s standard genetic distances (DS).

Figure 4.
Scatter plot showing the first, second, and third principal components of genetic differentiation among nine Cashmere goat populations.
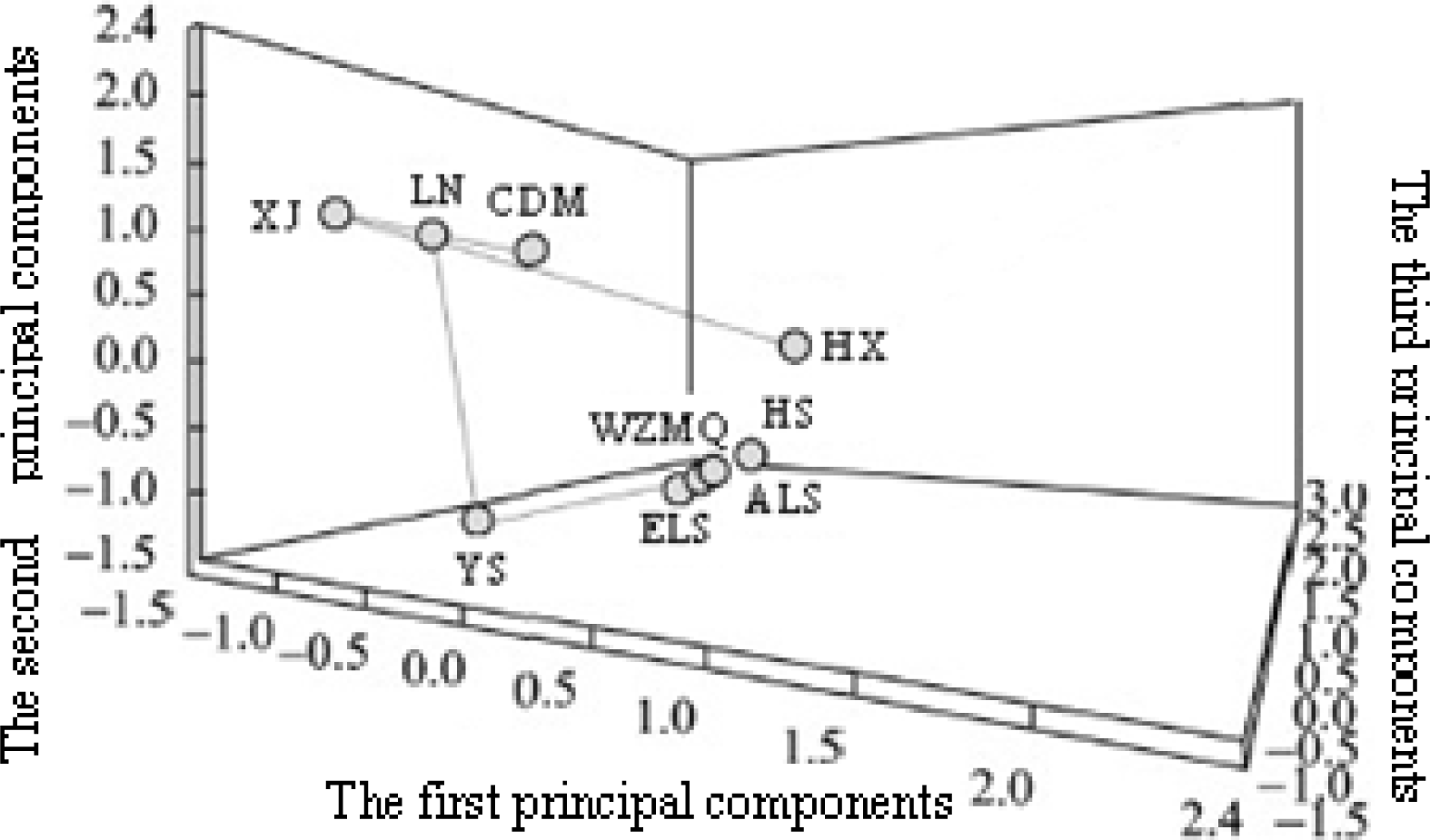
Table 1.
Name, code, sample size, and source region of nine Chinese cashmere goat populations
Table 2.
Information of 20 microsatellite loci
Table 3.
Basic genetic variation information of the nine Chinese Cashmere goat populations
Table 4.
F-statistics of nine Chinese cashmere goat populations at 20 microsatellite loci
| Locus |
All studied Chinese Cashmere goat populations
|
||||
|---|---|---|---|---|---|
| FIT | FST | FIS | GST | Nm | |
| OarFCB20 | 0.0785 | 0.0866** | 0.0088 | 0.0762 | 2.6368 |
| ILSTS029 | 0.1738** | 0.1029*** | 0.0790** | 0.1103 | 2.1795 |
| MAF70 | 0.0676 | 0.0758** | −0.0089 | 0.0608 | 3.0482 |
| MCM527 | 0.0738 | 0.0629** | 0.0116 | 0.0331 | 3.7246 |
| ILSTS087 | 0.1679** | 0.1215*** | 0.0528** | 0.1118 | 1.8076 |
| SPS113 | 0.1193* | 0.1054*** | 0.0155 | 0.0836 | 2.1219 |
| ILSTS005 | 0.1036* | 0.1372*** | −0.0389 | 0.0273 | 1.5722 |
| ILSTS011 | 0.1151* | 0.1385*** | −0.0272 | 0.1482 | 1.5551 |
| SRCRSP3 | 0.2421*** | 0.1973*** | 0.0558** | 0.1738 | 1.0171 |
| CSRD247 | 0.0309 | 0.0716** | −0.0438 | 0.0573 | 3.2416 |
| INRA063 | 0.1280* | 0.1469*** | −0.0222 | 0.1461 | 1.4518 |
| MAF065 | 0.0627 | 0.0915** | −0.0317 | 0.0690 | 2.4822 |
| OarFCB48 | 0.0806 | 0.0981** | −0.0194 | 0.0673 | 2.2984 |
| MAF209 | 0.2534*** | 0.1632*** | 0.1078*** | 0.1554 | 1.2819 |
| BMS1494 | 0.1986*** | 0.1358*** | 0.0727** | 0.1412 | 1.5909 |
| SRCRSP5 | 0.1782** | 0.1083*** | 0.0779** | 0.0427 | 2.0584 |
| BM1818 | 0.1548** | 0.1278*** | 0.0310* | 0.0538 | 1.7062 |
| DRBP1 | 0.2186*** | 0.1064*** | 0.1256*** | 0.0862 | 2.0996 |
| OarFCB304 | 0.1028* | 0.1369*** | −0.0395 | 0.0956 | 1.5762 |
| OarAE54 | 0.1968*** | 0.1535*** | 0.0512** | 0.1406 | 1.3787 |
| Total | 0.1374** | 0.1184*** | 0.0229* | 0.0940 | 2.0415 |
FIT: Wright’s F-statistics of inbreeding within total; FST: Wright’s F-statistics of subpopulation within total; FIS: Wright’s F-statistics of inbreeding within subpopulation; GST: Genetic differentiation coefficient; Nm: Gene flow; Nm = 0.25(1−FST)/FST. Significance levels of deficit in heterozygotes:
Table 5.
Nei′s genetic distances (shaded area above the diagonal) and Nei′s standard genetic distances (below the diagonal) among nine Chinese Cashmere goat populations
REFERENCES
Araújo AM, Guimaraes SEF, Machado TMM, Lopes PS, Pereira CS, Silva FLR, Rodrigues MT, Columbiano VS, Fonseca CG. 2006. Genetic diversity between herds of Alpine and Saanen dairy goats and naturalized Brazilian Moxotó breed. Genet Mol Biol 29:67–74.

Arranz J, Bayon Y, San Primitivo F. 1998. Genetic relationships among Spanish sheep using microsatellites. Anim Genet 29:435–440.


Avise JC. 2000. The history and formation of species. Cambridge: Harvard University Press, USA; p. 439
Balloux F, Lugon-Moulin N. 2002. The estimation of population differentiation with microsatellite markers. Mol Ecol 11:155–165.


Barker JSF, Tan SG, Moore SS, Mukherjee TK, Matheson JL, Selvaraj OS. 2001. Genetic variation within and relationships among populations of Asian goats (Capra hircus). J Anim Breed Genet 118:213–233.

Botstein D, White RL, Skolnick M, Davis RW. 1980. Construction of a genetic linkage map in man using restriction fragment polymorphisms. Am J Hum Genet 32:314–331.


Bowcock AM, Ruiz Linare A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. 1994. High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457.


Bruford MW, Wayne RK. 1993. Microsatellites and their application to population genetic studies. Curr Opin Genet Dev 3:939–943.


Bruno-de-Sousa C, Martinez AM, Ginja C, Santos-Silva F, Carolino MI, Delgado JV, Gama LT. 2011. Genetic diversity and population structure in Portuguese goat breeds. Livest Sci 135:131–139.

Canon J, García D, García-Atance MA, Obexer-Ruff G, Lenstra JA, Ajmone-Marsan P, Dunner S. 2006. Geographical partitioning of goat diversity in Europe and the Middle East. Anim Genet 37:327–334.


Chen SL, Fan B, Liu B, Yu M, Zhao SH, Zhu MJ, Xiong TG, Li K. 2006. Genetic variations of 13 indigenous Chinese goat breeds based on cytochrome b gene sequences. Biochem Genet 44:89–99.


Crawford AM, Dodds KG, Ede AJ, Pierson CA, Montgomery GW, Garmonsway HG, Beattie AE, Davies K, Maddox JF, Kappes SW, Stone RT, Nguyen TC, Penty JM, Lord EA, Broom JE, Buitkamp J, Schwaiger W, Epplen JT, Matthew P, Matthews ME, Hulme DJ, Beh KJ, McGraw RA, Beattie CW. 1995. An autosomal genetic linkage map of the sheep genome. Genetics 140:703–724.




Davis MB, Shaw RG. 2001. Range shifts and adaptive responses to quaternary climate change. Science 292:673–679.


Di R, Farhad Vahidi SM, Ma YH, He XH, Zhao QJ, Han JL, Guan WJ, Chu MX, Sun W, Pu YP. 2011. Microsatellite analysis revealed genetic diversity and population structure among Chinese cashmere goats. Anim Genet 42:428–431.


Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620.


Falush D, Stephens M, Pritchard KJ. 2003. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164:1567–1587.




Fatima S, Bhonga CD, Ranka DN, Joshi CG. 2008. Genetic variability and bottleneck studies in Zalawadi, Gohilwadi and Surti goat breeds of Gujarat (India) using microsatellites. Small Rumin Res 77:58–64.

Fernandez H, Hughes S, Vigne JD, Helmer D, Hodgins G, Miquel C, Hanni C, Luikart G, Taberlet P. 2006. Divergent mtDNA lineages of goats in an Early Neolithic site, far from the initial domestication areas. Proc Natl Acad Sci USA 103:15375–15379.



Gamaa LT, Bressan MC. 2011. Biotechnology applications for the sustainable management of goat genetic resources. Small Rumin Res 98:133–146.

Ganai NA, Yadav BT. 2001. Genetic variation within and among three Indian breeds of goat using heterologous microsatellite markers. Anim Biotechnol 12:121–136.


Glowatzki-Mullis ML, Muntwyler J, Baumle E, Gaillard C. 2008. Genetic diversity measures of Swiss goat breeds as decision making support for conservation policy. Small Rumin Res 74:202–211.

Goudet J. 2002. Fstat: a program to estimate and test gene diversities and fixation indices. Version 2.9.3.2. Available at http://www.unil.ch/izea/softwares/fstat.html
Gour DS, Malik G, Ahlawat SPS, Pandey AK, Sharma R, Gupta N, Gupta SC, Bisen PS, Kumar D. 2006. Analysis of genetic structure of Jamunapari goats by microsatellite markers. Small Rumin Res 66:140–149.

Groeneveld LF, Lenstra JA, Eding H, Toro MA, Scherf B, Pilling D, Negrini R, Finlay EK, Jianlin H, Groeneveld E, Weigend S; The GLOBALDIV Consortium. 2010. Genetic diversity in farm animals-a review. Anim Genet 41:6–31.


Guo X, Elston RC. 1999. Linkage informative content of polymorphic genetic markers. Hum Hered 49:112–118.


Gutierrez-Espeleta GA, Kalinowski ST, Boyce WM, Hedrick PW. 2000. Genetic variation and population structure in desert bighorn sheep: implications for conservation. Conserv Genet 1:3–15.

Hedrick PW. 1999. Highly variable loci and their interpretation in evolution and conservation. Evolution 53:313–318.


Hochberg Y. 1988. A sharper Bonferroni procedure for multiple test of significance. Biometrika 75:800–802.

Iamartino D, Bruzzone A, Lanza A, Blasi M, Pilla F. 2005. Genetic diversity of Southern Italian goat populations assessed by microsatellite markers. Small Rumin Res 57:249–255.

Kang JF, Li XL, Zhou RY, Li LH, Zheng GR, Zhao HY. 2011. Genetic diversity and differentiation of four goat lineages based on analysis of complete mtDNA d-loop. Front Agric In China 5:87–93.

Kantanen J, Olsaker I, Holm LE, Lien S, Vilkki J, Brusgaard K. 2000. Genetic diversity and population structure of 20 North European cattle breeds. J Hered 91:446–457.


Kimura M, Crow JF. 1964. The number of alleles that can be maintained in a finite population. Genetics 49:725–738.




Li MH, Zhao SH, Bian C, Wang HS, Wei H, Liu B, Yu M, Fan B, Chen SL, Zhu MJ, Li SJ, Xiong TA, Li K. 2002. Genetic relationships among twelve Chinese indigenous goat populations based on microsatellite analysis. Genet Sel Evol 34:729–744.



Li XL, Valentini A. 2004. Genetic diversity of Chinese indigenous goat breeds based on microsatellite markers. J Anim Breed Genet 121:350–355.

Luikart G, Gielly L, Excoffier L, Vigne JD, Bouvet J, Taberlet P. 2001. Multiple maternal origins and weak phylogeographic structure in domestic goats. Proc Natl Acad Sci USA 98:5927–5932.



MacHugh DE, Shiriver MD, Loftus RT, Cunningham P, Bradley DG. 1997. Microsatellite DNA variation and the evolution, domestication and phylogeography of Taurine and Zebu cattle (Bos taurus and Bos indicus). Genetics 146:1071–1086.




Martínez AM, Acosta J, Vega-Pla JL, Delgado JV. 2006. Analysis of the genetic structure of the canary goat populations using microsatellites. Livest Sci 102:140–145.

Martinez-Martinez A, Vega-Pla JL, Ribeiro MN, Camacho ME, Revidatti MA, Galarza A, Stemmer A, Sponenberg P, Seguí B, Gómez M, Martín D, Rocha LL, Chacón E, Pimenta E, Capote J, Amills M, Cabello A, Gama LT, Delgado JV, Menezes MPC. 2008. Genetic relationships between native goat populations from Spain, Portugal and America. In : Proceedings of the XXXI Conference of the International Society for Animal Genetics; Amsterdam, The Netherlands.
Muema EK, Wakhungu JW, Hanotte O, Jianlin H. 2009. Genetic diversity and relationship of indigenous goats of Sub-saharan Africa using microsatellite DNA markers. Livest Res Rural Dev 21.
Nei M. 1991. Relative efficiencies of different tree-making methods for molecular data. Phylogenetic Analysis of DNA Sequence. Miyamoto MM, Cracrsft J, editorsNew York: Oxford University Press; p. 90–128.

Nei M, Tajima F, Tateno Y. 1983. Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170.


Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590.




Oliveira JCV, Ribeiro MN, Rocha LL, Gomes-Filho MA, Delgado JV, Martinez AM, Menezes MPC, Bettencourt CM, Gama LT. 2010. Genetic relationships between two homologous goat breeds from Portugal and Brazil assessed by microsatellite markers. Small Rumin Res 93:79–87.

Oliveira JD, Igarashi ML, Machado TM, Miretti MM, Ferro JA, Contel EP. 2007. Structure and genetic relationships between Brazilian naturalized and exotic purebred goat domestic goat (Capra hircus) breeds based on microsatellites. Genet Mol Biol 30:356–363.

Ozgecan KA, Okan E. 2012. Assessment of genetic diversity, genetic relationship and bottleneck using microsatellites in some native Turkish goat breeds. Small Rumin Res 105:53–60.

Peter C, Bruford M, Perez T, Dalamitra S, Hewitt G, Erhardt G. 2007. Genetic diversity and subdivision of 57 European and Middle-Eastern sheep breeds. Anim Genet 38:37–44.


Pritchard KJ, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959.




Qi JF. 2004. Report on domestic animal genetic resources in China. China Agric. Press; Beijing:
Qi Y, Luo J, Han XF, Zhu YZ, Chen C, Liu JX, Sheng HJ. 2009. Genetic diversity and relationships of 10 Chinese goat breeds in the middle and Western China. Small Rumin Res 82:88–93.

Reynolds J, Weir BS, Cockerham CC. 1983. Estimation of the coancestry coefficient: basis for a shortterm genetics distance. Genetics 105:767–769.


Ruane J. 1999. A critical review of the value of genetic distance studies in conservation of animal genetic resources. J Anim Breed Genet 116:317–323.

Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425.

Sechi T, Usai MG, Casu S, Carta A. 2005. Genetic diversity of Sardinian goat population based on microsatellites. Ital J Anim Sci 4:58–60.

Slatkin M, Barton NH. 1989. A comparison of three indirect methods of estimating average levels of gene flow. Evolution 43:1349–1368.


Taberlet P, Valentini A, Rezaei HR, Naderi S, Pompanon F, Negrini R, Ajmone-Marsan P. 2008. Are cattle, sheep, and goats endangered species? Mol Ecol 17:275–284.


Tadlaoui-Ouafi A, Babilliot JM, Leroux C, Martin P. 2002. Genetic diversity of the two main Moroccan goat breeds: phylogenetic relationships with four breeds reared in France. Small Rumin Res 45:225–233.

Takezaki N, Nei M. 1996. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389–399.




Takezaki N, Nei M, Tamura K. 2010. POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol Biol Evol 27:747–752.



Toro MA, Fernández J, Caballero A. 2009. Molecular characterization of breeds and its use in conservation. Livest Sci 120:174–195.

Traoré A, álvarez I, Tambourá HH, Fernández I, Kaboré A, Royo LJ, Gutiérrez JP, Sangaré M, Ouédraogo-Sanou G, Toguyeni A, Sawadogo L, Goyache F. 2009. Genetic characterisation of Burkina Faso goats using microsatellite polymorphism. Livest Sci 123:322–328.

Tu Y, Jiang Y, Han ZY, Feng WQ. 1989. Sheep and Goat Breeds in China. Shanghai Scientific & Technical Publishers; Shanghai, China:
Visser C, Hefer CA, van Marle-Koter E, Kotze A. 2004. Genetic variation of three commercial and three indigenous goat populations in South Africa. S Afr J Anim Sci 34:24–27.
Wang J, Chen YL, Wang XL, Yang ZX. 2008. The genetic diversity of seven indigenous Chinese goat breeds. Small Rumin Res 74:231–237.

Wang PJ, Shi BL, Li YC, Wang JY, Bai HW, Jin SQ, Zhang FQ. 2008. Study on improvement of Xinjiang goat using Liaoning cashmere goat. Chin J Anim Sci 27:8–11.
Wilson GA, Rannala B. 2003. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191.




Wimmers K, Ponsuksili S, Hardgeg T, Valle-Zarate A, Mathur P, Horst P. 2000. Genetic distinctness of African, Asian and south American local chickens. Anim Genet 31:159–165.


Wright S. 1969. Evolution and the genetics of populations: the theory of gene frequencies. 2:Univ. Chicago Press; Chicago:
Wright S. 1978. Evolution and the genetics of populations: variability within and among natural populations. 4:University of Chicago press; Chicago, IL, USA:
Yeh F, Yang RC, Boyle T. 1997. Popgene, the user friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Albert; Edmonton:
- TOOLS





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





