 |
 |
Abstract
In Erhualian and Yorkshire reciprocal cross F1 pig populations, we examined the mRNA expression characteristic of liver-derived IGF-1, IGF-1R, IGF-2, IGF-2R and IGFBP-3 during the embryonic and postnatal developmental periods (E50, E70, E90, D1, D20, D70, D120 and D180). Our results demonstrated that the IGF-system genes mRNA levels exhibited an ontogenetic expression pattern, which was potentially associated with the porcine embryonic development, postnatal growth, organogenesis and even the initiation and acceleration of puberty. The expression pattern of IGF-system genes showed variation in the reciprocal cross (F1 YE and EY pigs). This study also involved the expression features of imprinted genes IGF-2 and IGF-2R. The parent-of-origin effect of imprinted genes was reflected by their differential expression between the reciprocal crosses populations. The correlation analysis also indicated that the regulatory network and mechanisms involved in the IGF system were a complex issue that needs to be more fully explored. A better understanding of IGF system components and their interactive mechanisms will enable researchers to gain insights not only into animal organogenesis but also into somatic growth development and even reproduction.
It has long been known that the insulin-like growth factors (IGFs) system represent a family, including two ligands (IGF-1 and IGF-2), two corresponding cell-surface receptors (IGF-1R and IGF-2R), and at least six high-affinity IGF-binding proteins (IGFBPs 1-6) that specifically bind IGF-1 and IGF-2. This complex system plays an essential role in normal human and animal development, including embryogenesis, pre- and postnatal growth and in the maintenance of tissue homeostasis (Baker et al., 1993; Clemmons, 1997; Annunziata et al., 2011; Li et al., 2011), while abnormal IGF-system components have been implicated in diverse cancer progression (Sachdev and Yee, 2001).
The liver is the major endocrine organ for IGF ligands and IGFBPs in the blood circulation. The IGF-1 and IGF-2 as well as the IGFBPs are delivered from the liver to IGF-responsive tissues by the circulation, to function as regulators of individual growth and development (LeRoith et al., 2001; Ryan and Goss, 2008; Velloso, 2008). A major role of liver-derived IGF-1 is to regulate GH secretion by a negative feedback loop pattern to inhibit pituitary GH secretion. Lack of liver-derived IGF-1 results in increased GH levels, in turn, GH can upregulate hepatic IGF-1 gene expression and then the serum IGF-1 levels (Tannenbaum et al., 1983; Sj├Čgren et al., 1999; Yakar et al., 1999; Christoforidis et al., 2005; Ohlsson et al., 2009). IGF-2 is also expressed in the liver, but is not as dependent on GH as IGF-1, especially in postnatal growth stage (Collett-Solberg and Cohen, 2000; Annunziata et al., 2011). Furthermore, The IGF-2 gene is normally paternally expressed in human and mice (Krassas et al., 2003; Haig, 2004) and has been suggested to affect development during preimplantation stages (Rappolee et al., 1992; Lighten et al., 1997). In pigs, the IGF-2 locus is particularly complex because of the tissue-specific imprinting and the IGF-2 isoforms-specific imprinting originating from different promoters, only some of which have been shown to be imprinted (Li et al., 2008). However, the porcine IGF-2 has been reported to be maternally imprinted in 10-wk-old fetal pig liver (Nezer et al., 1999). At present, IGF-2 is considered as a primary growth factor required for embryonic and fetal growth, while IGF-1 is required for achieving maximal postnatal growth (DeChiara et al., 1990; Gerrard et al., 1998). In the blood circulation, the majority of IGFs exist in a ternary complex with IGFBPs and the acid labile subunit (ALS). By binding IGFs, IGFBPs serve as circulating reservoirs transporting the IGFs, prolonging their half-life, and regulating their bioavailability and activity (JŪ┐rgensen, 2003). IGFBP-3 is the most abundant and predominant circulating IGFBP, which binds the majority of endogenous IGFs, especially for IGF-1 (Moses et al., 1979; Clemmons, 1997). In normal mice, 70 to 80% of IGF-1 exists as a ternary complex of IGF-1GFBP3-ALS in the circulation (LeRoith, 2008). On the other hand, the physiological actions of IGFs are mediated mainly by membrane receptors, the IGF-1R and IGF-2R, to guide downstream signaling transduction and gene activity (Cheng et al., 2002; Pavelic et al., 2007). The receptors are mainly expressed in IGFs target tissues (Adams et al., 2000). Liver cells also have IGFs receptors, mainly IGF-2 receptors (Froesch et al., 1985). In animal models, the breast and prostate growth were restrained after the disruption of the IGF-1R signaling pathway (Ruan et al., 1999), and knockout of the IGF-2R gene or loss of the imprinted IGF-2R showed fetal overgrowth and perinatal lethality due to major cardiac abnormalities (Lau et al., 1994; Wang et al., 1994), both suggested that IGFs receptors are involved in normal organogenesis in animals. At the same time, the IGF-2R gene is paternally imprinted in rodents, artiodactyls and marsupials, but is biallelically expressed in primates (Killian et al., 2000). In pigs, it has been shown that IGF-2R is maternally expressed (Killian et al., 2001; Bischoff et al., 2009; Cha et al., 2010).
As mentioned above, altered gene actions of the liver-derived IGFs system components have been implicated in the mediation of both pre- and postnatal growth of animals. But till now, little is known about the expression profiles of the IGFs components with different development stages in the liver tissues, so one objective of this study was to assess the mRNA expression of IGF component genes during the embryonic and postnatal development period. Moreover, it is unknown whether there are different expression patterns in reciprocal crosses. Offspring produced by a reciprocal cross are also an important genetic source material for studies on gene activity and even individual phenotypes, especially for imprinted genes. Both IGF-2 and IGF-2R are imprinted expression in pigs, but no data is available on their expression characteristics in porcine liver, let alone at specific developmental stages. So here, we investigate the differential expression of these genes in F1 hybrids from reciprocal crosses between Chinese native Erhualian and Western Yorkshire pigs to detect the effects of the reciprocal crosses on IGF-system genes expression, and the parent-of-origin effects on imprinted gene expression. At the same time, we analyze the relationship between changes in gene expression and porcine body weight, and examine the effects of liver-derived IGFs system on porcine growth and development in the reciprocal cross population.
Chinese native Erhualian and Western Yorkshire pigs were obtained from the breeding pig farm of Nanjing Hejia-jisheng agriculture and animal husbandry Co., Ltd. All animals were fed maintenance diets for standard growth under the same conditions. The reciprocal crosses were performed to produce F1 crossbreds from Yorkshire boars├ŚErhualian sows (F1: YE) and Erhualian boars├ŚYorkshire sows (F1: EY). The F1 hybrids from reciprocal crosses were used as experimental animals. Both in reciprocal YE and EY population, we randomly selected F1 crossbreds at 50 (E50), 70 (E70), and 90 (E90) days of embryonic stage and at 1 (D1), 20 (D20), 70 (D70), 120 (D120), and 180 (D180) days of postnatal development with 6 biological repeats (3 boars and 3 sows) at each stage. The liver tissues for each individual were collected after slaughter and immediately stored in liquid nitrogen until analysis. All protocols involving the use of pigs received prior approval from the Chinese Animal Care and Use Committee. The PrimeScriptŌäó RT reagent Kit, SYBR Premix TaqŌäó Kit, and Trizol were purchased from TaKaRa Bio Inc. (Dalian, China). All other chemicals were of reagent grade and were obtained from standard commercial sources.
Total RNA was extracted from the liver tissue using the Trizol reagent kit (Invitrogen, Carlsbad, CA) according to the manufacturerŌĆÖs instructions. RNA concentration was determined by the ultraviolet colorimetry method (OD260/OD280), and RNA integrity was evaluated by formaldehyde denatured agarose gel electrophoresis. The single-strand cDNA was synthesized by an PrimeScriptŌäó RT reagent Kit (Takara), following the protocol suggested by the manufacturer. The RT reaction was performed at 37┬░C for 15 min followed by 85┬░C for 5 s. The synthesized cDNA solutions were diluted 5-fold and then stored at ŌłÆ20┬░C before real-time quantitative RT-PCR.
For detecting the expression level of IGFs system components, i.e. IGF-1, IGF-2, IGF-1R, IGF-2R, and IGFBP-3 genes, real-time quantitative PCR (qRT-PCR) was performed using SYBR┬« Premix Ex TaqŌäó (Takara, Dalian, China) according to the manufacturerŌĆÖs instructions. Glyceraldeyhyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. Primers were designed based on the porcine mRNA sequences from the GenBank database for all these genes, using the Premier 5 software (PREMIER Biosoft Int., Palo Alto, CA). All primers were synthesized by Invitrogen (Shanghai, China). Each PCR reaction mixture (20-╬╝l) consisted of 10 ╬╝l SYBR┬« Premix Ex TaqŌäó, 0.4 ╬╝l forward primer, 0.4 ╬╝l reverse primer, 7.2 ╬╝l sterile water, and 2.0 ╬╝l cDNA solution. The following protocol was used: one cycle of 5 min at 95; 40 cycles of 10 s at 95┬░C, 10 s at the annealing temperature of the primers (Table 1), 15 s at 72┬░C, plate-reading; 72┬░C for 10 min, followed by plate-reading every other 0.2┬░C from 65┬░C to 94┬░C for drawing melting curves; 72┬░C for 10 min; then the reaction was ended with a maintain at 4┬░C. Amplification and melt curve analysis were performed using a thermocycler (Opticon 2, MJ Research, Waltham, MA, USA). Following the manufacturerŌĆÖs instruction, the expression level of each gene was analyzed according to previously described methods (Lu et al., 2008; Lu et al., 2010). For each gene, controls for each primer set containing no cDNA were included on each plate, and the reaction was repeated three times for every sample on each plate. The amplification profiles of each gene are shown in Table 1.
Data were described as mean┬▒SEM and statistically analyzed using SPSS 17.0 for windows statistical package (SPSS Inc., Chicago, IL, USA). The level of statistical significance was 0.05. After normalized by GAPDH, the differences in IGF-system genes expression in different developmental stages were analyzed by ANOVA process, followed by the Tukey-Kramer test as a multiple comparison test. The t-test was performed to compare gene expression differences between reciprocal cross pigs at the same developmental stage. The relationships among the mRNA expression of IGF-system genes, and the association between changes in gene expression and porcine body weight, were examined by calculating the Pearson correlation coefficient.
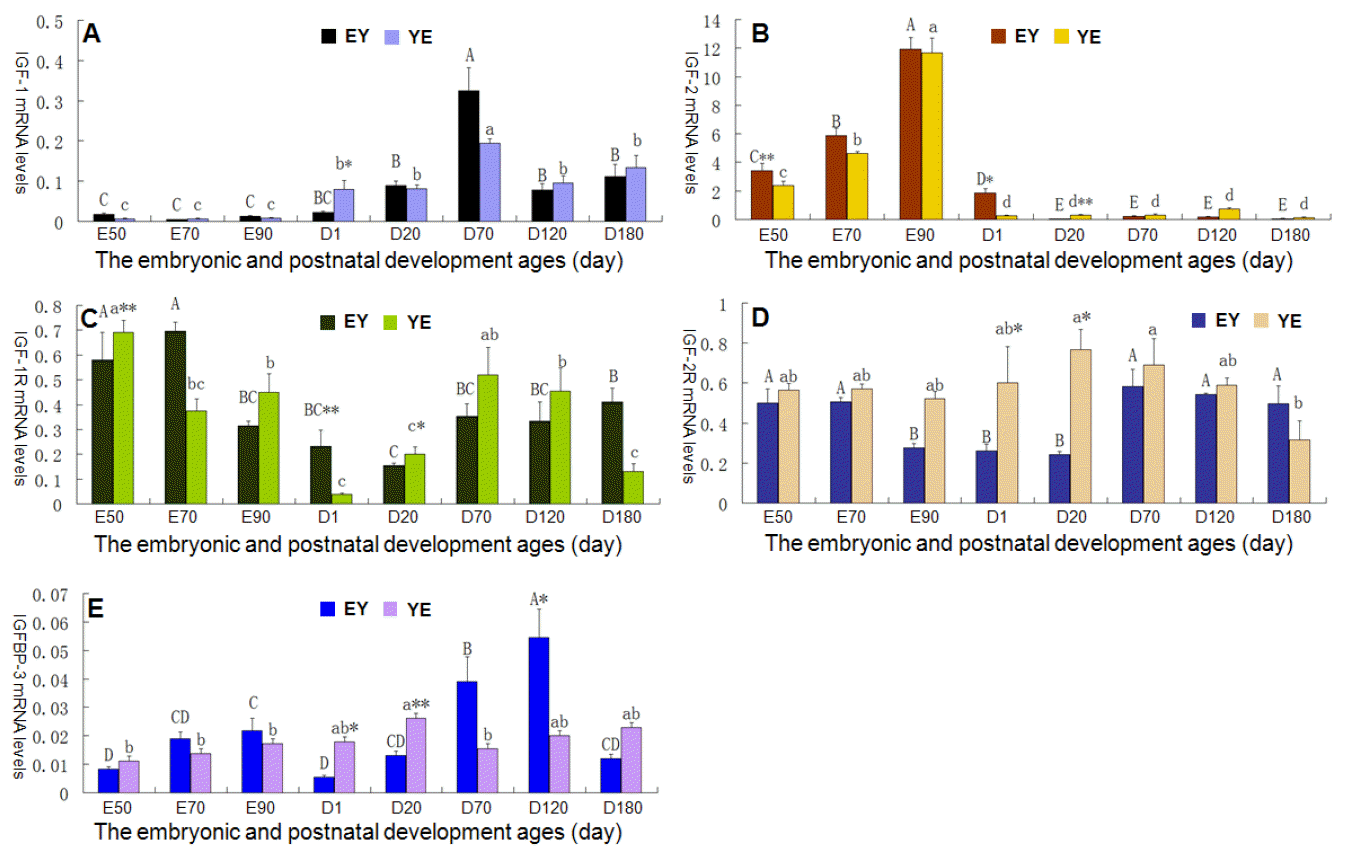
The RNAs and cDNAs all meet the quality control for qRT-PCR analysis (showed in supplemental materials). The amplification products of all the primers had single bright bands with the expected sizes, and the melting-curve showed only one peak. Figure 1 indicates the expression profiles of IGF-system component genes during the embryonic and postnatal developmental stages in the liver tissue of YE and EY pigs.
The changes in IGF-1 mRNA expression are shown in Figure 1A. In F1 crossbreds from EY pigs, IGF-1 transcripts were almost not detected in the early development stage from the age E50 to the period around birth (D1), and there were no significant expression differences among the ages E50, E70, E90, and D1. However after D20, IGF-1 expression rapidly increased, and reached a peak value at 70 days old, then the mRNA expression decreased significantly with the individuals development until D120 (p<0.01). From age D120 to D180, the IGF-1 transcripts maintained at a relative low level, but its mRNA expression was much higher than that of the early embryonic period (p<0.01). In F1 crossbreds from YE pigs, there was also a distinct expression characteristic for IGF-1 gene between the embryonic periods and the postnatal developmental stages in the porcine liver. IGF-1 transcripts in the embryonic period were remarkably lower than that of any stages after the birth (p<0.05 or p<0.01). IGF-1 mRNA firstly remained at a stable low level during the embryonic growth, then dramatically increased to the higher level at D1 (p<0.05) after birth. At around age D70, IGF-1 mRNA expression reached peak levels, and then markedly decreased to a lower level at age D120 (p<0.05) but maintained at a low level from age D120 to D180. At the same time, by comparing the IGF-1 mRNA expression difference between EY and YE F1 crossbreds at a same developmental stage, it was indicated that there was only one distinct mRNA level at age D1 (p<0.05), while at other times no significant differences were found.
The IGF-2 mRNA expression patterns are shown in Figure 1B. The relatively abundant IGF-2 mRNAs presented in fetal life of EY F1 pigs. With the development of embryo, IGF-2 mRNA level increased continuously from age E50, and then reached a peak at E90. A sharp decline in IGF-2 mRNA expression occurred at postnatal day 1 (p<0.01) and then a significant decrease again appeared at D20 (p<0.05). From age D20 to D180, IGF-2 mRNA level remained at a relative low level. The IGF-2 mRNA expression pattern in F1 YE crossbreds was similar to that in EY crossbreds, except that the mRNA level had dropped to a very low level in YE crossbreds just at age D1, but not D20. For EY and YE F1 crossbreds at a same developmental stage, t-test revealed that the expression of IGF-2 mRNA was higher in EY F1 pigs compared with YE F1 pigs at age E50 (p<0.01) and at D1 (p<0.05), whereas the opposite was true at D20 (p<0.01).
Figure 1C indicates the changes in IGF-1R mRNA expression levels. IGF-2R showed a high expression level at age E50 and E70 in EY pigs and then significantly decreased through E90 and D1 (p<0.01). At around D20, IGF-2R mRNA dropped to the lowest level, and then its mRNA expression gradually increased till D180 to reach a peak level. This was markedly higher than that at age D20 (p<0.01), while lower than that at E50 (p<0.05). In YE F1 pigs, we found the expression pattern or tendency of IGF-1R mRNA was similar with that in EY pigs. However, IGF-1R mRNA levels significantly decreased (p<0.01) at age E70 compared with E50, and at around age D1 fallen to the lowest value (p<0.01). Then IGF-1R mRNA increased continuously through age D20 and reached a peak at D70 (p<0.01), following by a drop in a stepwise fashion through D120 and D180. On the other hand, IGF-1R mRNA expression level was higher in YE pigs compared with EY pigs at age E50 (p<0.05) and D20 (p<0.01), and conversely, the EY pigs had the higher IGF-1R level at age D1 (p<0.01).
The changes in IGF-2R mRNA expression are shown in Figure 1D. In EY F1 pigs, IGF-2R mRNA remained at high levels on ages E50 and E70, then dramatically decreased at E90 (p<0.05) and hereafter maintained at a low level till D20. A significant increase in IGF-2R mRNA was observed at age D70 (p<0.01), and it maintained at a relatively stable high level until D180. There was no significant IGF-2R mRNA expression difference among ages E50, E70, D70, D120, and D180. In YE F1 pigs, a marked IGF-2R mRNA difference presented on ages D20 (p<0.01) and D70 (p<0.01) when comparing with D180, while there was no significant difference among the other developmental stages. Meantime, the expression of IGF-2R mRNA was higher in F1 EY pigs compared with YE pigs at ages D1 and D20 (p<0.05).
The IGFBP-3 mRNA expression profiles are shown in Figure 1E. IGFBP-3 mRNA was expressed at a low level during the embryonic periods in EY F1 pigs, with a progressive increase from E50 to E90 (p<0.05). A significant decrease in IGFBP-3 mRNA occurred at D1 (p<0.05), then it gradually increased from age D20 and reached a peak level at D120, and then a marked decrease emerged at age D180 in EY F1 pigs (p<0.01). In YE F1 pigs, IGFBP-3 also showed a low expression during fetal life. There was no distinct IGFBP-3 mRNA expression among ages E50, E70, E90, and D1. After birth, IGFBP-3 mRNA level weakly increased and reached a peak value at age D20, and then a significant decrease appeared at D70 (p<0.05) and afterwards it remained at a relatively stable level until D180. The difference analysis between reciprocal cross populations showed that: the expression of IGFBP-3 mRNA was higher in F1 YE pigs than that in EY pigs at ages D1 (p<0.05) and D20 (p<0.01), while the contrary was true at age D120 (p<0.01).
The IGF-1 binds IGF-1R with high affinity, and IGF-2 interacts with IGF-1R and IGF-2R. The IGFBP-3 has key roles in regulating ligand bioavailability by binding IGF-1 or IGF-2. So in our study, the relationships among the mRNA expression of IGF-system genes were analyzed based on different developmental stages. We picked out the significant correlation items in EY or YE F1 pigs as shown in Table 2. In EY F1 pigs, IGF-1 mRNA level was positively correlated with IGF-2R and IGFBP-3 both in E50-D180 and D1-D180, while negatively correlated with IGF-2 in E50-D180. Also a positive correlation between IGF-1R and IGF-2R, and between IGF-2R and IGFBP-3 existed in both E50-D180 and D1-D180. In YE F1 pigs, there was a positive correlation between IGF-1 and IGFBP-3 during the embryonic periods, and between IGF-2 and IGFBP-3. IGF-1 mRNA level was also positively correlated with IGF-2R from age E50 to D180.
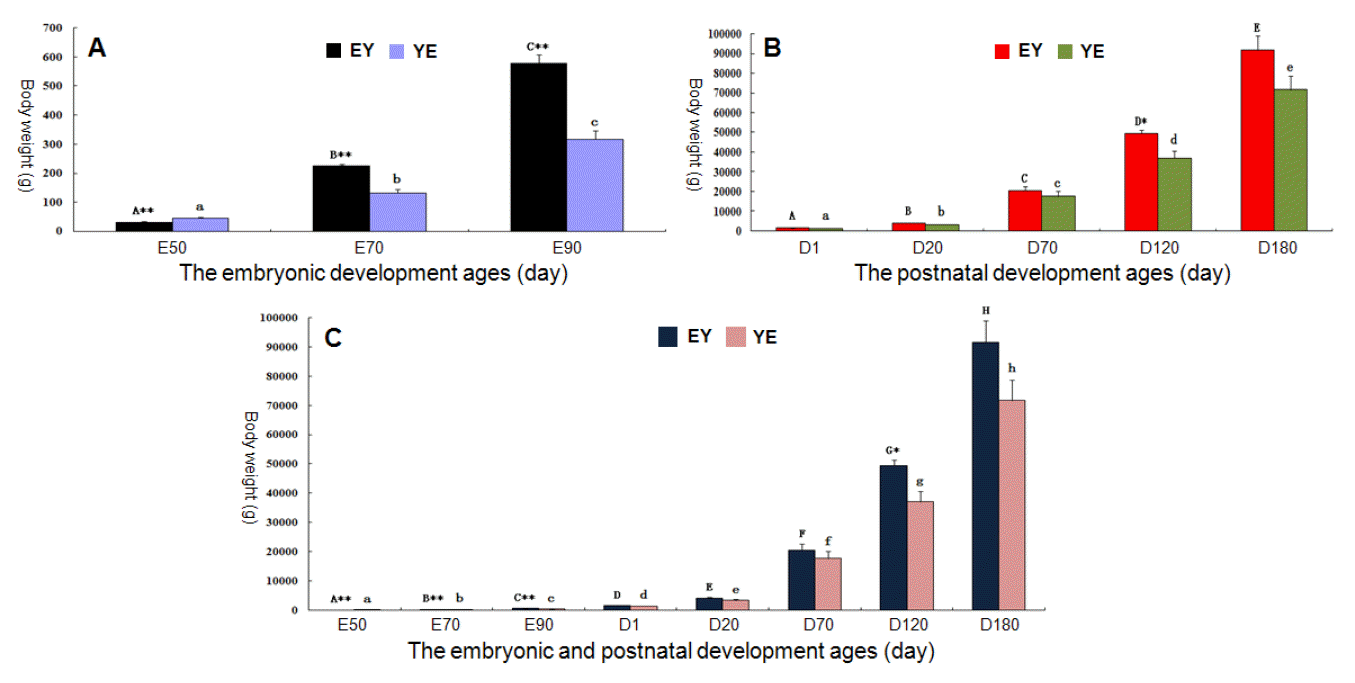
Figure 2 shows the changes in body weight during the embryonic and postnatal developmental stages in EY and YE F1 pigs. There were significant body-weight differences among the developmental periods E50, E70, E90, D1, D20, D70, D120 and D180 both in EY and YE populations (p<0.01). The body weight of EY F1 pigs was markedly higher than that of YE pigs in the same developmental stage during the embryonic period (p<0.01) (Figure 2A), while during the postnatal developmental stages, no remarkable difference existed except for age D120 (p<0.05) (Figure 2B).
The associations between the mRNA expression of IGF-system genes and body weight were analyzed based on different developmental stages in EY or YE F1 pigs, and the significant correlation items are shown in Table 3. Both in EY and YE F1 pigs, the body weights were positively correlated with IGF-2 and IGFBP-3 during the embryonic stages, and negatively correlated with IGF-2R in the same time. IGF-2 and IGF-2R were observed to be negatively and positively correlated with body weight during the postnatal developmental periods (D1-D180) in EY F1 pigs respectively, but not in YE pigs. In addition, IGF-1 and IGFBP-3 were positively correlated with body weight in YE F1 pigs from age E50 to D180, but not in EY pigs.
Over the past decades, numerous studies have indicated that IGF-1 and its receptor IGF-1R play an essential role in animal growth development and reproductive function. IGF-1 null mice had marked growth retardation in utero and postnatal periods. These mice had abnormal body weight both at birth and the peri-pubertal growth stage, and they were also infertile (Baker et al., 1993; Powell-Braxton et al., 1993; Liu et al., 1998). In many species, including human, studies have demonstrated that serum levels of IGF-1 were associated with the initiation and acceleration of puberty (Hiney et al., 1991; Laron and Klinger, 1998). After puberty, serum IGF-1 levels progressively declined (Argente et al., 1993). The Erhualian boars and sows reach their puberty at around 70 and 60 days of age respectively, and later for the Yorkshire pigs (Li et al., 2003). Our results further confirmed the potential effects of liver-derived IGF-1 on animal reproduction during postnatal development, because both EY and YE F1 pigs had a relatively high IGF-1 mRNA levels at about age D70, and similarly after that, the IGF-1 levels markedly decreased. Simultaneously, IGF-1 is a critical component for growth stimulation. Studies had provided evidence that IGF-1 plays an important role in body growth in dogs, i.e. low serum IGF-1 associated with low body weight (Eigenmann et al., 1988; Tryfonidou et al., 2003). Skalkidou et al. (2003) also reported that IGF-1 plays a dominant role in pre- and perinatal growth in human. However, most studies supported that IGF-1 played a larger role in neonatal and postnatal growth (Heyner et al., 1990; De Pablo et al., 1991; Baker et al., 1993; Liu et al., 1998). In this study, a progressive increase pattern of IGF-1 level along with porcine growth and development also indicated an important role of IGF-1 for individual growth. In addition, there was a positive correlation between IGF-1 mRNA levels and porcine body weight from age E50 to D180 in the YE F1 population. During the embryonic periods, IGF-1 was expressed at relatively low levels. There were no significant changes among ages E50, E70, and E90. However after birth, IGF-1 expression markedly stepped up. A previous study also showed a low level expression of IGF-1 during the embryonic period, which was considered more important for postnatal growth and development (Powell-Braxton et al., 1993). Furthermore, animals with liver-specific IGF-1 knockout showed similar body weight to controls at birth, although they had only 25% or less serum IGF-1 levels compared with controls (Sj├Čgren et al., 1999; Yakar et al., 1999), while genetically elevated liver IGF-1 expression resulted in increased postnatal body growth in mice (Stratikopoulos et al., 2008). Our results are consistent with the standpoint of neonatal and postnatal stimulation function for serum IGF-1, at least in pigs. On the other hand, there was a significant mRNA expression difference between EY and YE F1 pigs at age D1, i.e. the rapid increase of IGF-1 level after birth in YE F1 pigs was earlier than that in EY pigs. This also hints a possibility that there may be a different time point for IGF-1 to exert its effect between different breeds, but it demands additional study to confirm. At the same time, IGF-1R was identified as an essential regulator of organogenesis, IGF1R-null mice died shortly after birth due to organ hypoplasia (Baker et al., 1993; Liu et al., 1993). IGF-1R conditional knockout in the liver decreased the capacity for liver regeneration (Desbois-Mouthon et al., 2006). In our study, a lowest IGF-1R level appeared in the perinatal periods. It may represent a relatively decreased activity of the liver organ at this stage. Moreover, no significant correlation between IGF-1R mRNA level and porcine body weight was found in our analysis. This still highlights the organogenetic function of liver-derived IGF-1R rather than the somatic growth and development. Additionally, a similar change tendency of IGF-1R mRNA expression existed in both EY and YE F1 pigs, but there were marked IGF-1R level differences between the YE and EY F1 pigs at ages E50, D20 and D1. This may be attributed to the time variance for organogenesis between individuals with different genetic background. However, the specific reason and mechanism remain to be elucidated.
IGF-2 is the most important fetal growth factor which shared biochemical and biological properties with IGF-1, while its receptor IGF-2R is believed to act as a negative regulator responsible for clearing excess IGF-2 during fetal development (Collett-Solberg and Cohen, 2000; Rabnott et al., 2003). Researchers suggested IGF-2 was more influential in embryonic development rather than postnatal somatic growth (DeChiara et al., 1990). IGF-2 null mice also showed growth impairment, but this occurred only in utero while their postnatal growth was normal (Baker et al., 1993; Powell-Braxton et al., 1993). Serum IGF-2 concentration was high in fetal and neonatal life, but it declined rapidly after birth (Moses et al., 1980). In this study, the relatively abundant IGF-2 mRNAs presented during the embryonic periods both in EY and YE F1 pigs, whereas in postnatal stages the opposite was true. After birth, the IGF-2 mRNAs sharply dropped to a very low level. Our results are consistent with previous studies, suggesting that IGF-2 plays a significant role during fetal development. Furthermore, our correlation analysis between gene expression and porcine body weight also supports this viewpoint, because a significant positive correlation existed during the fetal life while a slight negative from age D1 to D180. The expression of IGF-2R mRNA was relatively stable except a slight drop during perinatal periods in EY F1 pigs. This may be associated with the expression changes of IGF-2, but it demands additional study to confirm. In addition, IGF-2R mRNA levels were negatively correlated with porcine body weight during embryonic periods, which was consistent with the role of IGF-2R as a negative regulator for fetal development (Collett-Solberg and Cohen, 2000; Rabnott et al., 2003). On the other hand, both IGF-2 and IGF-2R are imprinted genes in pigs. IGF-2 has been reported to be paternally expressed, while IGF-2R is maternally expressed (Nezer et al., 1999; Killian et al., 2001; Bischoff et al., 2009; Cha et al., 2010). If a gene is imprinted, the relative allelic expression activity and even individual phenotype may be different in progeny of reciprocal crosses (Chaillet 1994; Curley and Keverne, 2004; Wittkopp et al., 2006). So in present study, we detected the parental allele-specific expression of imprinted IGF-2 and IGF-2R by using F1 offspring from reciprocal crosses. It was found that there were expression differences between EY and YE F1 pigs during the early embryonic period and early postnatal growth stages, and after birth the IGF-2 mRNA level in YE F1 pigs decreased faster or earlier than that in EY pigs. The male parent was Yorkshire boars for YE pigs, and Erhualian boars for EY pigs. Because of maternal imprinting, IGF-2 is expressed only from the paternal allele. It is possible that these differences are related to parent-of-origin effects on imprinted gene expression within the two breeds. As for the porcine body weight phenotype, there was significant difference between EY and YE F1 pigs in the same developmental stage during the embryonic periods. This was consistent with the standpoint proposed by Haig and Graham (1991), suggesting that imprinting of growth factors such as IGF-2, IGF-2R regulated embryonic growth in the mammalian uterus (Haig and Graham, 1991).
IGFBPs function as carrier proteins and regulators in the circulation by binding IGFs. Many studies showed that IGFBP-3 was the most dominant IGFBP in postnatal serum, binding to IGFs with the highest affinity (Jone and Clemmons, 1995; Levitt Katz et al., 1995; Rajaram et al., 1997). In our study, the expression of IGFBP-3 mRNA exhibited a fluctuant pattern and its levels did not change acutely in YE F1 pigs, but in EY pigs IGFBP-3 levels markedly fluctuated, reaching its low peak at perinatal period and high peak value at around puberty respectively. These variances indicated a complex physiological role for liver-derived IGFBP-3, but the underlying mechanism needs to be revealed by in-depth study. A previous study has shown showed that IGFBP-3 has growth-promoting and inhibiting effects both in vivo and in vitro (Collett-Solberg and Cohen, 2000). In addition, there was a complicated relationship among IGFBPs and IGFs. The IGFBPs could modulate IGF activity, transport and increase IGF half-life. Simultaneously, they could also inhibit IGFs receptor action by competitive binding with IGF ligands (Annunziata et al., 2011; Jones and Clemmons, 1995). On the contrary, IGFs could also influence IGFBP activity. For example, IGF-1 up-regulated IGFBP-3 at the transcriptional and/or posttranscriptional levels (Bale and Conover, 1992). In vitro study showed a production of IGFBP-3 in liver cells after IGF-1 stimulation (Uchijima et al., 1995). This was consistent with our result, indicating that IGF-1 mRNA level was positively correlated with IGFBP-3 both in E50-D180 and D1-D180. Moreover, we also found some other significant correlations among the IGF-system component genes. In fact, as a complex regulatory network for animal growth and development, the IGFs system exerts its biological functions not only within the liver, but also involves the extra-liver tissues. Studies have demonstrated that it might influence hypothalamic and pituitary processes to facilitate growth and development, such as the GH/IGF-1 axis (Lackey et al., 1999). Additionally, locally produced (extra-liver tissues) IGF system components may also play an important role for normal growth and development. Of course, species difference may be another important factor which needs to be considered. In a word, as more and more related researches continue to be carried out when considering more comprehensive factors, the exact physiological role and regulatory mechanism of IGF system component genes will become better elucidated.
In conclusion, in Erhualian and Yorkshire reciprocal cross F1 pig populations, we examined the mRNA expression characteristic of liver-derived IGF-1, IGF-1R, IGF-2, IGF-2R and IGFBP-3 during the embryonic and postnatal developmental periods. Our results demonstrated that the IGF-system gene mRNA levels exhibited an ontogenetic expression pattern, which was potentially associated with the porcine embryonic development, postnatal growth, organogenesis and even the initiation and acceleration of puberty. This study also involved the expression features of imprinted genes IGF-2 and IGF-2R in pigs. The parent-of-origin effect of imprinted genes was reflected by their differential expression between the reciprocal crosses populations. This is the first study that reports changes in the relative abundance of mRNAs for IGF-system components throughout the embryonic and postnatal development in the reciprocal cross F1 pigs. However, the regulatory network and mechanisms involved in the IGF system are a complex issue that needs to be more fully explored. With a better understanding of IGF system components and their interactive mechanisms, researchers will be able to gain insights not only into animal organogenesis but also into somatic growth development and even reproduction property.
ACKNOWLEDGEMENTS
This work was supported by the Chinese National Programmes for High Technology Research and Development (863 Programme, No. 2007AA10Z149), the National Transgenic Major Project (No. 2008ZX08006-003), and the Natural Science Foundation of Jiangsu Province (No. BK2011650).
Figure┬Ā1
The expression profiles of IGF-system component genes in the porcine liver during the embryonic and postnatal developmental stages. A, B, C, D and E represent the mRNA expression profiles of IGF-1, IGF-2, IGF-1R, IGF-2R and IGFBP-3 respectively. YE = Yorkshire boar├ŚErhualian sow crossbreds; EY = Erhualian boar├ŚYorkshire sow crossbreds. Different letters denotes statistically significant differences among developmental stages in one breed. The capital letter represents EY pigs and the lower case letter YE pigs. The symbol * indicates a significant difference between F1 EY and YE pigs at the same age with the significance level p<0.05, and ** indicates a significant difference with p<0.01.
Figure┬Ā2
The changes in porcine body weight during the embryonic and postnatal developmental stages. YE = Yorkshire boar├ŚErhualian sow crossbreds; EY = Erhualian boar├ŚYorkshire sow crossbreds. Different letters denote statistically significant differences among developmental stages in one breed. The capital letter represents EY pigs and the lower case letter YE pigs. The symbol * indicates a significant difference between F1 EY and YE pigs at the same age with the significance level p<0.05, and * indicates a significant difference with p<0.01.
Table┬Ā1
Primer sequences and PCR condition for detection of mRNA
| Gene name | Accession No. | Primer sequence (5ŌĆ▓ŌåÆ3ŌĆ▓) | Size (bp) | AT1 (┬░C) |
|---|---|---|---|---|
| GAPDH | AF017079 |
F2: GGACTCATGACCACAGTCCAT R2: TCAGGTCCACAACCGACACGT |
220 | 57 |
| IGF-1 | DQ784687 |
F: ATTTCTTGAAGGTAAAGATGCA R: CAGCCCCACAGAGGGTCTCA |
117 | 59 |
| IGF-2 | NM213883 |
F: CCCAGTGAGACTCTGTGCG R: CAGGTGTCATAGCGGAAGAAC |
275 | 57 |
| IGF-1R | AB003362 |
F: CGAGAGACATCTATGAGACA R: TCCTCACTGTAGTAGAAGGA |
382 | 57 |
| IGF-2R | AF339885 |
F: ATCCTCAATCCCATAGCC R: CTCTTACAATGAAACGCAAT |
110 | 50 |
| IGFBP-3 | AF085482 |
F: GACACGCTGAACCACCTCA R: CGTACTTATCCACGCACCAG |
151 | 57 |
Table┬Ā2
Correlation analysis of IGF-system genes mRNA level during the embryonic and postnatal developmental period in EY and YE F1 pigs
| Gene 1 | Gene 2 | Period |
Pearson correlation
|
|
|---|---|---|---|---|
| EY | YE | |||
| IGF-1 | IGF-2 | E50-D180 | ŌłÆ0.347* (p = 0.013, N = 51) | ŌłÆ0.282 (p = 0.055, N = 47) |
| IGF-2R | E50-D180 | 0.317* (p = 0.024, N = 51) | 0.311* (p = 0.034, N = 47) | |
| IGF-2R | D1-D180 | 0.371* (p = 0.04, N = 31) | 0.178 (p = 0.365, N = 28) | |
| IGFBP-3 | E50-D180 | 0.539** (p = 0, N = 51) | 0.048 (p = 0.747, N = 47) | |
| IGFBP-3 | E50-E90 | ŌłÆ0.151 (p = 0.524, N = 20) | 0.480* (p = 0.037, N = 19) | |
| IGFBP-3 | D1-D180 | 0.554** (p = 0.001, N = 31) | ŌłÆ0.158 (p = 0.421, N = 28) | |
| IGF-1R | IGF-2R | E50-D180 | 0.356* (p = 0.01, N = 51) | ŌłÆ0.136 (p = 0.362, N = 47) |
| IGF-2R | D1-D180 | 0.452* (p = 0.011, N = 31) | ŌłÆ0.16 (p = 0.415, N = 28) | |
| IGF-2 | IGFBP-3 | E50-E90 | 0.435 (p = 0.055, N = 20) | 0.582** (p = 0.009, N = 19) |
| IGF-2R | IGFBP-3 | E50-D180 | 0.385** (p = 0.005, N = 51) | ŌłÆ0.04 (p = 0.791, N = 47) |
| IGFBP-3 | D1-D180 | 0.496** (p = 0.005, N = 31) | ŌłÆ0.212 (p = 0.279, N = 28) | |
Table┬Ā3
Correlation between IGF-system genes mRNA level and porcine body weight during the embryonic and postnatal developmental period in EY and YE F1 pigs
| Genes | Period |
Pearson correlation
|
||
|---|---|---|---|---|
| EY | YE | |||
| Body weight | IGF-1 | E50-D180 | 0.256 (p = 0.07, N = 51) | 0.387** (p = 0.007, N = 47) |
| IGF-2 | E50-E90 | 0.519* (p = 0.019, N = 20) | 0.761** (p = 0.000, N = 19) | |
| D1-D180 | ŌłÆ0.414* (p = 0.021, N = 31) | ŌłÆ0.246 (p = 0.207, N = 28) | ||
| IGF-2R | E50-E90 | ŌłÆ0.576** (p = 0.008, N = 20) | ŌłÆ0.636** (p = 0.003, N = 19) | |
| D1-D180 | 0.409* (p = 0.022, N = 31) | ŌłÆ0.037 (p = 0.853, N = 28) | ||
| IGFBP-3 | E50-D180 | 0.199 (p = 0.161, N = 51) | 0.304* (p = 0.038, N = 47) | |
| E50-E90 | 0.538* (p = 0.015, N = 20) | 0.523* (p = 0.022, N = 19) | ||
REFERENCES
Adams T, Epa V, Garrett T, Ward C. 2000. Structure and function of the type 1 insulin-like growth factor receptor. Cell Mol Life Sci 57:1050ŌĆō1093.


Argente J, Barrios V, Pozo J, Munoz M, Hervas F, Stene M, Hernandez M. 1993. Normative data for insulin-like growth factors (IGFs), IGF-binding proteins, and growth hormone-binding protein in a healthy Spanish pediatric population: age-and sex-related changes. J Clin Endocrinol Metab 77:1522ŌĆō1528.


Baker J, Liu JP, Robertson EJ, Efstratiadis A. 1993. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75:73ŌĆō82.


Bale LK, Conover CA. 1992. Regulation of insulin-like growth factor binding protein-3 messenger ribonucleic acid expression by insulin-like growth factor I. Endocrinology 131:608ŌĆō614.


Bischoff SR, Tsai S, Hardison N, Motsinger-Reif AA, Freking BA, Nonneman D, Rohrer G, Piedrahita JA. 2009. Characterization of conserved and nonconserved imprinted genes in swine. Biol Reprod 81:906ŌĆō920.



Cha BH, Kim BK, Hwang S, Yang BC, Im GS, Park MR, Woo JS, Kim MJ, Seong HH, Cho JH. 2010. Imprinted gene mRNA expression during porcine peri-implantation development. Asian-Aust J Anim Sci 23:693ŌĆō699.

Chaillet JR. 1994. Genomic imprinting: lessons from mouse transgenes. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 307:441ŌĆō449.

Cheng R, Chang KM, Wu JL. 2002. Different temporal expressions of Tilapia (oreochromis mossambicus) insulin-like growth factor-I and IGF binding protein-3 after growth hormone induction. Mar Biotechnol 4:218ŌĆō225.

Christoforidis A, Maniadaki I, Stanhope R. 2005. Growth hormone/insulin-like growth factor-1 axis during puberty. Pediatr Endocrinol Rev 3:5ŌĆō10.

Clemmons DR. 1997. Insulin-like growth factor binding proteins and their role in controlling IGF actions. Cytokine Growth Factor Rev 8:45ŌĆō62.


Collett-Solberg P, Cohen P. 2000. Genetics, chemistry, and function of the IGF/IGFBP system. Endocrine 12:121ŌĆō136.


Curley JP, Keverne EB. 2004. Imprinting and behavior Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics. John Wiley & Sons, Ltd.
De Pablo F, Robcis HL, Caldes T, Alemany J, Scavo L, Serrano J. 1991. Insulin-like growth factor-I and insulin as growth and differentiation factors in chicken embryogenesis. Poult Sci 70:1790ŌĆō1796.


DeChiara TM, Efstratiadis A, Robertsen EJ. 1990. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345:78ŌĆō80.


Desbois-Mouthon C, Wendum D, Cadoret A, Rey C, Leneuve P, Blaise A, Housset C, Tronche F, Le Bouc Y, Holzenberger M. 2006. Hepatocyte proliferation during liver regeneration is impaired in mice with liver-specific IGF-1R knockout. FASEB J 20:773ŌĆō775.


Eigenmann JE, Amador A, Patterson DF. 1988. Insulin-like growth factor I levels in proportionate dogs, chondrodystrophic dogs and in giant dogs. Acta Endocrinol (Copenh) 118:105ŌĆō108.


Froesch E, Schmid C, Schwander J, Zapf J. 1985. Actions of insulin-like growth factors. Annu Rev Physiol 47:443ŌĆō467.


Gerrard D, Okamura C, Ranalletta M, Grant A. 1998. Developmental expression and location of IGF-I and IGF-II mRNA and protein in skeletal muscle. J Anim Sci 76:1004ŌĆō1011.


Haig D. 2004. Genomic imprinting and kinship: how good is the evidence? Annu Rev Genet 38:553ŌĆō585.


Haig D, Graham C. 1991. Genomic imprinting and the strange case of the insulin-like growth factor II receptor. Cell 64:1045ŌĆō1046.


Heyner S, Farber M, Rosenblum I. 1990. The insulin family of peptides in early mammalian development. Curr Top Dev Biol 24:137ŌĆō159.


Hiney JK, Ojeda S, Dees WL. 1991. Insulin-like growth factor I: a possible metabolic signal involved in the regulation of female puberty. Neuroendocrinology 54:420ŌĆō423.


JŪ┐rgensen HL. 2003. Serum levels of insulin-like growth factor I and its binding proteins in health and disease. Ugeskr L Ger 165:3429
Jones JI, Clemmons DR. 1995. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev 16:3ŌĆō34.


Killian JK, Byrd JC, Jirtle JV, Munday BL, Stoskopf MK, MacDonald RG, Jirtle RL. 2000. M6P/IGF2R imprinting evolution in mammals. Mol Cell 5:707ŌĆō716.


Killian JK, Nolan CM, Wylie AA, Li T, Vu TH, Hoffman AR, Jirtle RL. 2001. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum Mol Genet 10:1721ŌĆō1728.


Krassas G, Pontikides N, Kaltsas T, Dumas A, Frystyk J, Chen J, Flyvbjerg A. 2003. Free and total insulin-like growth factor (IGF)-I,-II, and IGF binding protein-1,-2, and-3 serum levels in patients with active thyroid eye disease. J Clin Endocrinol Metab 88:132ŌĆō135.


Lackey BR, Gray SL, Henricks DM. 1999. The insulin-like growth factor (IGF) system and gonadotropin regulation: actions and interactions. Cytokine Growth Factor Rev 10:201ŌĆō217.


Laron Z, Klinger B. 1998. Effect of insulin-like growth factor-I treatment on serum androgens and testicular and penile size in males with Laron syndrome (primary growth hormone resistance). Eur J Endocrinol 138:176ŌĆō180.


Lau M, Stewart C, Liu Z, Bhatt H, Rotwein P, Stewart CL. 1994. Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev 8:2953ŌĆō2963.


Le Roith D, Bondy C, Yakar S, Liu JL, Butler A. 2001. The somatomedin hypothesis: 2001. Endocr Rev 22:53ŌĆō74.


LeRoith D. 2008. Clinical relevance of systemic and local IGF-I: lessons from animal models. Pediatr Endocrinol Rev 5:739ŌĆō743.

Levitt Katz LE, Rosenfeld RG, Cohen SP. 1995. Clinical significance of insulin-like growth factor binding proteins (IGFBPs). Endocrinologist 5:36ŌĆō43.

Li C, Bin Y, Curchoe C, Yang L, Feng D, Jiang Q, Tian XC, Zhang S. 2008. Genetic imprinting of H19 and IGF2 in domestic pigs (Sus scrofa). Anim Biotechnol 19:22ŌĆō27.


Li F, Zhao R, Xu Q, Chen W, Ma Y, Chen J. 2003. Characteristics of testosterone secretion in male Erhualian and Large White pigs in different developmental stages. J Nanjing Agricultural University 26:117ŌĆō119.
Li S, Yakar S, Brodt P. 2011. Role of the IGF-axis in liver metastasis: experimental and clinical evidence. Brodt P, editorLiver metastasis: biology and clinical management. Springer; Netherlands: p. 233ŌĆō271.

Lighten AD, Hardy K, Winston RML, Moore GE. 1997. IGF2 is parentally imprinted in human preimplantation embryos. Nat Genet 15:122ŌĆō123.


Liu JL, Grinberg A, Westphal H, Sauer B, Accili D, Karas M, LeRoith D. 1998. Insulin-like growth factor-I affects perinatal lethality and postnatal development in a gene dosage-dependent manner: manipulation using the Cre/loxP system in transgenic mice. Mol Endocrinol 12:1452ŌĆō1462.


Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. 1993. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (IGF1R). Cell 75:59ŌĆō72.


Lu F, Jiang Z, Wang X, Luo Y, Li X, Liu H. 2010. Role of the insulin-like growth factor system in epiphyseal cartilage on the development of Langshan and Arbor Acres chickens, Gallus domesticus. Poult Sci 89:956ŌĆō965.


Lu F, Wang X, Pan Q, Huang R, Liu H. 2008. Expression of genes involved in the somatotropic, thyrotropic, and corticotropic axes during development of Langshan and Arbor Acres chickens. Poult Sci 87:2087ŌĆō2097.


Moses AC, Nissley PS, Passamani J, White RM. 1979. Further characterization of growth hormone-dependent somatomedin-binding proteins in rat serum and demonstration of somatomedin-binding proteins produced by rat liver cells in culture. Endocrinology 104:536ŌĆō546.


Moses AC, Nissley SP, Short PA, Rechler MM, White RM, Knight AB, Higa OZ. 1980. Increased levels of multiplication-stimulating activity, an insulin-like growth factor, in fetal rat serum. Proc Natl Acad Sci USA 77:3649ŌĆō3653.



Nezer C, Moreau L, Brouwers B, Coppieters W, Detilleux J, Hanset R, Karim L, Kvasz A, Leroy P, Georges M. 1999. An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat Genet 21:155ŌĆō156.


Ohlsson C, Mohan S, Sj├Čgren K, Tivesten , Isgaard J, Isaksson O, Jansson JO, Svensson J. 2009. The role of liver-derived insulin-like growth factor-I. Endocr Rev 30:494ŌĆō535.



Pavelic J, Radakovic B, Pavelic K. 2007. Insulin-like growth factor 2 and its receptors (IGF 1R and IGF 2R/mannose 6-phosphate) in endometrial adenocarcinoma. Gynecol Oncol 105:727ŌĆō735.


Powell-Braxton L, Hollingshead P, Warburton C, Dowd M, Pitts-Meek S, Dalton D, Gillett N, Stewart TA. 1993. IGF-I is required for normal embryonic growth in mice. Genes Dev 7:2609ŌĆō2617.


Rabnott G, Kaye P, Pantaleon M, ericho H. 2003. The role of insulin-like growth factor II and its receptor in mouse preimplantation development. Reprod Fertil Dev 15:37ŌĆō45.


Rajaram S, Baylink DJ, Mohan S. 1997. Insulin-like growth factor-binding proteins in serum and other biological fluids: regulation and functions. Endocr Rev 18:801ŌĆō831.


Rappolee DA, Sturm KS, Behrendtsen O, Schultz GA, Pedersen RA, Werb Z. 1992. Insulin-like growth factor II acts through an endogenous growth pathway regulated by imprinting in early mouse embryos. Genes Dev 6:939ŌĆō952.


Ruan W, Powell-Braxton L, Kopchick JJ, Kleinberg DL. 1999. Evidence that insulin-like growth factor I and growth hormone are required for prostate gland development. Endocrinology 140:1984ŌĆō1989.


Ryan PD, Goss PE. 2008. The emerging role of the insulin-like growth factor pathway as a therapeutic target in cancer. Oncologist 13:16ŌĆō24.

Sj├Čgren K, Liu JL, Blad K, Skrtic S, Vidal O, Wallenius V, LeRoith D, T rnell J, Isaksson OGP, Jansson JO. 1999. Liver-derived insulin-like growth factor I (IGF-I) is the principal source of IGF-I in blood but is not required for postnatal body growth in mice. Proc Natl Acad Sci USA 96:7088ŌĆō7092.



Skalkidou A, Petridou E, Papathoma E, Salvanos H, Kedikoglou S, Chrousos G, Trichopoulos D. 2003. Determinants and consequences of major insulin-like growth factor components among full-term healthy neonates. Cancer Epidemiol Biomarkers Prev 12:860ŌĆō865.

Stratikopoulos E, Szabolcs M, Dragatsis I, Klinakis A, Efstratiadis A. 2008. The hormonal action of IGF1 in postnatal mouse growth. Proc Natl Acad Sci USA 105:19378ŌĆō19383.



Tannenbaum G, Guyda HJ, Posner BI. 1983. Insulin-like growth factors: a role in growth hormone negative feedback and body weight regulation via brain. Science 220:77ŌĆō79.


Tryfonidou M, Holl M, Vastenburg M, Oosterlaken-Dijksterhuis M, Birkenh ger-Frenkel D, Van den Brom W, Hazewinkel H. 2003. Hormonal regulation of calcium homeostasis in two breeds of dogs during growth at different rates. J Anim Sci 81:1568ŌĆō1580.


Uchijima Y, Takenaka A, Takahashi SI, Noguchi T. 1995. Production of insulin-like growth factors and their binding proteins in primary cultures of rat liver parenchymal and nonparenchymal cells. Biosci Biotechnol Biochem 59:1503ŌĆō1515.


Velloso CP. 2008. Regulation of muscle mass by growth hormone and IGF-I. Br. J Pharmacol 154:557ŌĆō568.



Wang ZQ, Fung MR, Barlow DP, Wagner EF. 1994. Regulation of embryonic growth and lysosomal targeting by the imprintedIGF2/Mpr gene. Nature 372:464ŌĆō467.







 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





