 |
 |
| Anim Biosci > Volume 31(10); 2018 > Article |
|
Abstract
Objective
In birds, three types of melatonin receptors (MTNR1A, MTNR1B, and MTNR1C) have been cloned. Previous researches have showed that three melatonin receptors played an essential role in reproduction and ovarian physiology. However, the association of polymorphisms of the three receptors with duck reproduction traits and egg quality traits is still unknown. In this test, we chose MTNR1A, MTNR1B, and MTNR1C as candidate genes to detect novel sequence polymorphism and analyze their association with egg production traits in Shaoxing duck, and detected their mRNA expression level in ovaries.
Methods
In this study, a total of 785 duck blood samples were collected to investigate the association of melatonin receptor genes with egg production traits and egg quality traits using a direct sequencing method. And 6 ducks representing two groups (3 of each) according to the age at first eggs (at 128 days of age or after 150 days of age) were carefully selected for quantitative real-time polymerase chain reaction.
Results
Seven novel polymorphisms (MTNR1A: g. 268C>T, MTNR1B: g. 41C>T, and g. 161T>C, MTNR1C: g. 10C>T, g. 24A>G, g. 108C>T, g. 363 T>C) were detected. The single nucleotide polymorphism (SNP) of MTNR1A (g. 268C>T) was significantly linked with the age at first egg (p<0.05). And a statistically significant association (p<0.05) was found between MTNR1C g.108 C>T and egg production traits: total egg numbers at 34 weeks old of age and age at first egg. In addition, the mRNA expression level of MTNR1A in ovary was significantly higher in late-mature group than in early-mature group, while MTNR1C showed a contrary tendency (p<0.05).
Melatonin (N-acetyl-5-methoxytryptamine) is an important hormone that is synthesized mainly in the pineal gland, and has a profound effect on serval physiology process including circadian rhythm and reproduction through its special receptors in birds [1–3]. Three melatonin receptor subtypes, MTNR1A (alias MT1, Mel1a), MTNR1B (alias MT2, Mel1b), and MTNR1C (alias Mel1c), which belong to the superfamily of G protein-coupled receptors [4], have been cloned in birds [5,6].
In chicken, melatonin receptor subtypes were identified in ovaries [7,8], suggesting that melatonin directly affects ovarian function through activating of multiple receptors. In geese, the expression levels of MTNR1A, MTNR1B, and MTNR1C initially increased and later decreased during follicular development cycle, indicating that melatonin receptors participated in activating small white follicles and small yellow follicles to develop into subsequent greater hierarchical follicles [9]. In addition, in situ hybridization of MTNR1C mRNA combined with immunocytochemistry for gonadotropin-inhibitory hormone (GnIH), a key neurohormone controlling avian reproduction by inhibiting gonadal development [10], revealed a clear cellular colocalization of MTNR1C mRNA and GnIH in paraventricular nuclei [11]. It seems that melatonin most likely acts directly on GnIH neurons through MTNR1C to induce GnIH expression and regulate avian reproduction.
Shaoxing duck is a Chinese dominant layer breed, characterized by small body size, early maturity and high productivity. Modern Shaoxing duck achieve 50% egg production by 140 days of age, and the mean number of eggs at 500 days old was more than 300 [12]. Shaoxing ducks have been considered as a good source of duck eggs in China, and improving its egg production performance is of primary interest to breeders and farmers. The identification of single nucleotide polymorphisms (SNP) in candidate genes associating with economically important traits has become a powerful tool for genetic improvement of animal selection and production.
Numerous studies have investigated the relationship between melatonin receptor subtypes and egg production traits in different species, and made melatonin receptor genes potential candidate genes for QTLs [13,14]. However, whether or not nucleotide polymorphisms of MTNR1A, MTNR1B, and MTNR1C are associated with the egg production traits and egg quality traits in ducks is still unknown. Therefore, this experiment aimed to detect the SNPs in MTNR1A, MTNR1B, and MTNR1C and explore their associations with egg production traits and egg quality traits. The expression levels of these genes in ovary of ducks at different age at first egg were also determined.
A total of 785 female Shaoxing ducks were randomly selected and raised in separate cages under similar environmental conditions and diet. The egg production traits of all ducks were recorded throughout the egg production process in terms of age at first egg, egg weight at 34 weeks old and total number of eggs at 34 weeks old and 72 weeks old. Egg collection for egg quality measurements took place at 34 weeks old. The following traits were recorded: egg shape index, shell thickness, shell strength, albumen height, Haugh unit score and eggshell color.
The blood samples of all the 785 individual ducks were collected from the wing vein using vacuum tubes containing dipotassium ethylene diamine tetraacetate (EDTA-K2) as an anticoagulant for further SNP analysis. To investigate the expression of the multiple forms of melatonin receptor mRNA, 6 ducks representing two groups (3 of each) according to the age at first egg (at 128 days of age or after 150 days of age) were carefully selected at age of 150 days. Ovaries were sampled after slaughter. These experiments were conducted in accordance with the Law of the People’s Republic of China on Animal Protection.
Based on the complete DNA sequence of Anas platyrhynchos genomic DNA sequence (NCBI accession no. NW_004676748.1), four pairs of primers were designed to amplify the target regions for SNP genotyping using the Primer Premier 6.0 software. Primer sequences are listed in Table 1.
Polymerase chain reaction (PCR) was carried out in a total volume of 15 μL consisting of 1 μL genomic DNA, 0.15 μL (10 μM) of each primer, 1.5 μL 10× buffer, 1.5 μL (25 mmol/L) MgCl2, 0.3 μL (10 mmol/L) dNTPs, and 1.5 U Taq DNA polymerase. PCR conditions were as follow: 95°C for 3 min, followed by 35 cycles of 95°C for 15 s, 55°C for 15 s, 72°C for 30 s, and a final extension of 72°C for 10 min. The PCR products were sequenced commercially using ABI 3730XL automated sequencer (Applied Biosystems, Carlsbad, CA, USA) after being purified and extended. SNPs were identified by looking for multiple peaks at the same base pair.
The genotypic frequencies were calculated for each individual and the Hardy-Weinberg equilibrium was analyzed using the Chi-square test of PopGene Version 1.32. Pairwise tests for linkage disequilibrium (LD) were performed for each SNP using the SHEsisPlus online software platform (http://shesisplus.bio-x.cn/SHEsis.html). The egg production traits were compared among the genotypes. The association between the SNPs and different traits in 785 ducks were analyzed using SPSS 22 with the model Y = μ+G+L+G×L+e, where Y is the dependent variable (analyzed traits), μ is the overall mean, G is the genotype with a variation for the candidate gene, L is the fixed effect of breed, G×L is the interaction between the genotype and duck population (a fixed effect), and e is the random error. The significance of the least squares means was tested with lest significance difference test.
Total RNA was extracted from ovary samples using Trizol reagent according to the manufacturer’s protocol. cDNA was synthesized from the total RNA according to the manufacturer’s protocol of TransScript First-Strand cDNA Synthesis SuperMix (TransGen, Beijing, China). Quantitative real-time PCR was performed using ABI 7500 (Applied Biosystems, Foster City, CA, USA). The mRNA expression levels were determined by Applied Biosystems real-time PCR using the MTNR1A, MTNR1B, and MTNR1C mRNA-specific primers and the β-acting gene as the internal control (Table 1). The 20 μL amplification reaction contained 10 μL of SYBR Green Universal PCR Master Mix, 2 μL cDNA, 0.4 μM of each primer and nuclease free water up to 20 μL. Thermal parameter used to amplify the template started with an initial denaturation at 94°C for 3 min followed by 40 cycles of 94°C for 10 s and annealing at 60°C for 30 s. The relative expression levels of the genes test were calculated using the 2−ΔCt method. Groups with different letters were significantly different at p<0.05.
Seven SNPs were identified in all three melatonin receptor gene exons respectively by direct sequencing (Figure 1), their corresponding allele and genotype frequencies are presented in Table 2. Hardy-Weinberg equilibrium tests showed that the alleles of MTNR1A g. 268C>T, MTNR1C g. 10C>T, and MTNR1C g. 108C>T were in Hardy-Weinberg equilibrium (p>0.05), and others deviated from the Hardy-Weinberg equilibrium (p<0.05) (Table 1). The LD tests of Shaoxing duck population showed that two SNPs (g. 41C>T, g. 161T>C) identified in MTNR1B gene were completely linked (D′ = 1.0 and r2 = 1.0) (Supplementary Table S1), providing only three possible haplotypes.
The association between the SNPs and egg production traits are shown in Table 3. For the MTNR1A g. 268C>T locus, age at first egg of the genotype CT was significantly less than that of the genotype TT (p<0.05). The g. 108C>T locus of MTNR1C was significantly associated with age at first egg and total number of eggs during 34-week egg-laying period (p<0.05). For MTNR1C g. 108C> T, the egg number of the genotype CT at 34 weeks old was significantly higher than that of the genotype CC (p<0.05), and ducks with the genotype TT exhibited significantly earlier age at first egg than that of the genotype CC (p<0.05). No significant association was found among other SNPs and egg weight, total number of eggs at 34 weeks old, total number of eggs at 72 weeks old or age at first egg (p>0.05).
As shown in Table 4, there were no significant association of 7 SNPs with egg quality trait. Furthermore, relation of SNPs with the color of eggshell was analyzed using chi-square test. When white eggshell was compared with blue eggshell, no significant differences in genotypes of MTNR1B or MTNR1C were observed except MTNR1A (p>0.05). The genotype TT and CT of g. 268C>T in MTNR1A were more common in Shaoxing ducks laying white eggshell compared with blue eggshell (Supplementary Table S2), suggesting MTNR1A g. 268C>T SNP may affect the eggshell color.
The expression of MTNR1A, MTNR1B, and MTNR1C between early-mature (first egg at 128 days of age) group and late-mature (first egg after 150 days of age) group showed different patterns (Figure 1). The late-mature group had significantly higher MTNR1A mRNA expression (0.0684±0.0017) compared to the early-mature group (0.0491±0.0060) (p<0.05) (Figure 2a), whereas there was no significant difference of MTNR1B mRNA expression between the early-mature group (1.491±0.4580) and the late-mature group (0.869±0.2435) (p> 0.05) (Figure 2b). The MTNR1C expression level of ovary was significantly higher in the early-mature group (2.4328±0.3936) than in late-mature group (p<0.05) (0.4756±0.0595) (Figure 2c).
Age at first egg, an important trait indicating sexual maturation and egg production performance, was negatively correlated with number of eggs [15–17]. However, age at first egg was controlled by polygenes [17] with low to moderate estimated heritability ranging from 0.13 to 0.20 [18–20], making the conventional breeding method ineffective. Therefore, molecular assisted selection becomes a powerful tool for improving egg production traits and raising economic benefits.
As previously reported in other avian species, melatonin regulates gonadal maturation by suppressing luteinizing hormone (LH) secretion, and stimulating GnIH and gonadotropin releasing hormone secretion [21–24]. MTNR1C mRNA colocalized with GnIH neurous [11] indicating that melatonin participates in gonadal maturation through binding its receptors. Therefore, melatonin receptors would be possible markers for selecting an early maturing breed. Although there were many studies seeking correlations between markers of candidate genes and age at first egg, such as growth hormone, prolactin, neuropeptide Y, follicle-stimulating hormone receptor, and SH3-domain GRB2-like 2 [25–29], little was known regarding polymorphisms in melatonin receptors of duck.
Our study demonstrated that g. 268C>T of MTNR1A gene shows a strong association with age of first egg. Beside, g. 268C>T of MTNR1C gene was found to be associated with total number of eggs at age of 34 weeks and age at first egg. A study in chickens also showed that SNPs (JQ249890:g. 384T>C, JQ249891:g. 387 T>C) locating at MTNR1A and SNP (JQ249896:g. 294 G>A) locating at MTNR1C were significantly linked with age at first egg [14]. The novel SNPs which was founded in our study, indicated that melatonin receptor genes may affect age at first egg and involve in sexual mature of ducks.
The exact role of melatonin receptors in different species during the reproductive process is not well understood. It was reported that melatonin receptor density showed a striking downregulation in brain when songbirds were less than fully reproductively mature, and subsequent resumed during photorefractory state [30]. While Abd et al [31] reported that MTNR1B and MTNR1C expression increased accompanying a delay in sexual maturity of Japanese quails. In our study, the expression of MTNR1C mRNA in late-mature group was significantly decreased compared to the early-mature group. Summarizing the results of the above researches, it is reasonable to suggest that MTNR1C activates avian sexual mature by down-regulating its expression level. The MTNR1A mRNA expression in our experiment showed a contrary tendency with MTNR1C mRNA, suggesting melatonin receptors regulate ovarian function through different mechanisms. Further studies are continuing to elucidate the exact role of melatonin receptors in avian sexual maturity.
Collectively, the results of present study suggest that the MTNR1A and MTNR1C genes play an important role in egg production of ducks, but also SNPs in these genes could be used as markers in molecular marker-assisted selection for duck reproduction traits.
ACKNOWLEDGMENTS
The authors acknowledge National Shaoxing Duck Conservation Farm staff for their technical work on the experiments. The authors would also like to thank the reviewers for helpful comments and insightful contributions that have led to the final version of this paper. This work was sponsored by the earmarked fund for National Waterfowl-industry Technology Research System (CARS-42-15 and CARS-42-06) and New Variety Breeding of Livestock and Poultry (2016C02054-12).
Figure 1
The C/T transition at base position 108 in the exon of MTNR1C gene. The box shows the single nucleotide polymorphism location. MTNR1C, melatonin receptor.
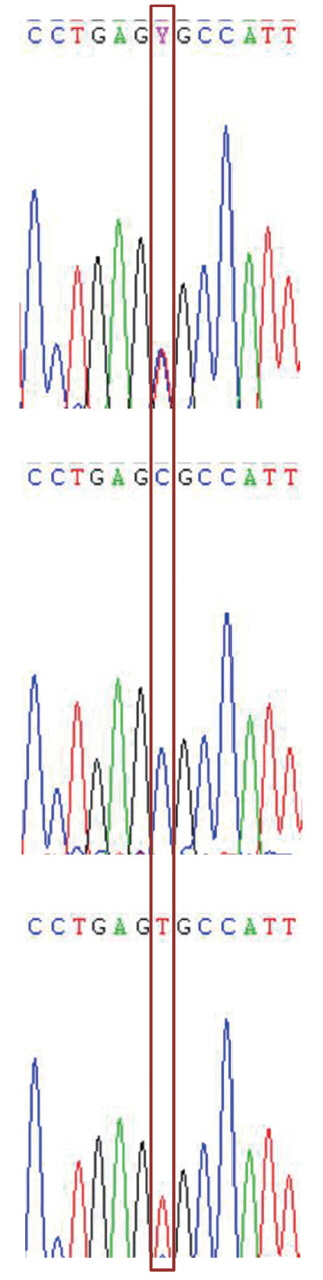
Figure 2
Results of expression of melatonin receptor subtypes mRNA in ovaries between early-mature group and late-mature group. (a) Comparison of mRNA expression of MTNR1A in ovaries between early-mature group and late-mature group. (b) Comparison of mRNA expression of MTNR1B in ovaries between early-mature group and late-mature group. (c) Comparison of mRNA expression of MTNR1C in ovaries between early-mature group and late-mature group. MTNR1A, MTNR1B, MTNR1C, melatonin receptor.
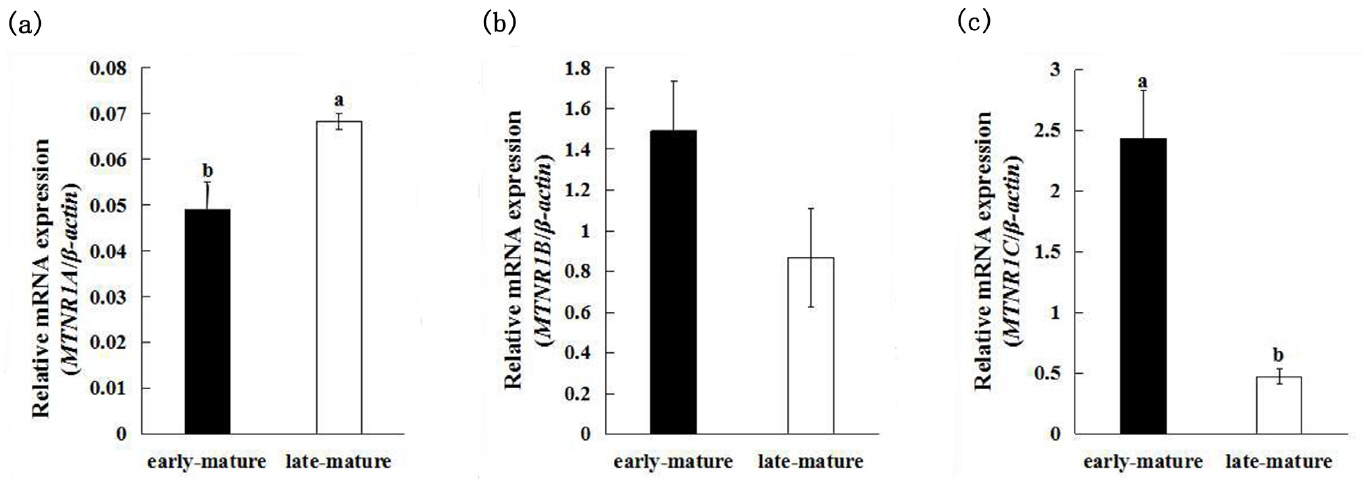
Table 1
Primers and sequencing information
Table 2
Genotypic and allelic frequency at the SNP loci of MTNR1A, MTNR1B, and MTNR1C genes in the Shaoxing duck population
| SNP | Genotype | No. of ducks | Genotype frequency | Allele | Allele frequency | χ2 |
|---|---|---|---|---|---|---|
| g. 268C>T | CC | 461 | 0.587 | C | 0.767 | 0.013 |
| (MTNR1A) | CT | 282 | 0.359 | T | 0.233 | |
| TT | 42 | 0.054 | ||||
| g. 41C>T | CC | 298 | 0.380 | C | 0.581 | 23.742* |
| (MTNR1B) | CT | 316 | 0.403 | T | 0.419 | |
| TT | 171 | 0.218 | ||||
| g. 161T>C | TT | 298 | 0.380 | T | 0.581 | 23.742* |
| (MTNR1B) | TC | 316 | 0.403 | C | 0.419 | |
| CC | 171 | 0.218 | ||||
| g. 10C>T | CC | 252 | 0.321 | C | 0.563 | 0.223 |
| (MTNR1C) | CT | 380 | 0.484 | T | 0.437 | |
| TT | 153 | 0.195 | ||||
| g. 24A>G | AA | 167 | 0.213 | A | 0.486 | 6.821* |
| (MTNR1C) | AG | 429 | 0.546 | G | 0.514 | |
| GG | 189 | 0.241 | ||||
| g. 108C>T | CC | 499 | 0.636 | C | 0.795 | 0.450 |
| (MTNR1C) | CT | 250 | 0.318 | T | 0.205 | |
| TT | 36 | 0.046 | ||||
| g. 363T>C | TT | 339 | 0.432 | T | 0.643 | 4.907* |
| (MTNR1C) | TC | 332 | 0.423 | C | 0.357 | |
| CC | 114 | 0.145 |
Table 3
Association between polymorphisms in MTNR1A, MTNR1B, and MTNR1C genes and egg production traits in Shaoxing ducks1)
| SNP | Genotype | Association of SNP with egg production traits | |||
|---|---|---|---|---|---|
|
|
|||||
| Egg weight | Number of eggs at 34 weeks of age | Number of eggs at 72 weeks of age | Age at first egg | ||
| g. 268C>T | CC | 70.45±5.17 | 75.99±5.00 | 303.23±12.93 | 146.51±15.18ab |
| (MTNR1A) | CT | 69.61±5.56 | 77.49±4.94 | 302.64±11.43 | 143.74±15.10b |
| TT | 68.57±8.20 | 74.95±3.73 | 298.19±15.49 | 146.55±15.30a | |
| g. 41C>T | CC | 70.13±5.46 | 76.64±4.87 | 303.84±15.23 | 146.21±15.77 |
| (MTNR1B) | CT | 70.33±5.59 | 75.88±4.98 | 304.28±16.49 | 145.85±14.87 |
| TT | 69.37±5.52 | 77.29±4.92 | 299.23±11.94 | 143.68±14.73 | |
| g. 161T>C | TT | 70.13±5.46 | 76.64±4.87 | 303.84±15.23 | 146.21±15.77 |
| (MTNR1B) | TC | 70.33±5.59 | 75.88±4.98 | 304.28±16.49 | 145.85±14.87 |
| CC | 69.37±5.52 | 77.29±4.92 | 299.23±11.94 | 143.68±14.73 | |
| g. 10C>T | CC | 69.98±5.59 | 75.53±4.81 | 303.49±13.99 | 145.95±14.44 |
| (MTNR1C) | CT | 69.92±5.57 | 77.23±4.79 | 306.12±14.44 | 145.26±15.73 |
| TT | 70.46±5.34 | 76.16±5.40 | 306.40±16.93 | 145.45±15.15 | |
| g. 24A>G | AA | 70.44±5.28 | 78.25±4.10 | 302.93±14.29 | 144.54±13.60 |
| (MTNR1C) | AG | 69.85±5.63 | 76.11±4.86 | 296.98±13.30 | 145.62±15.41 |
| GG | 70.13±5.51 | 75.72±5.68 | 304.29±17.11 | 146.16±16.06 | |
| g. 108C>T | CC | 70.25±5.73 | 75.09±5.34b | 298.91±11.83 | 146.50±15.75a |
| (MTNR1C) | CT | 69.79±5.24 | 78.91±3.76a | 302.65±14.23 | 144.28±14.28ab |
| TT | 68.98±4.56 | 78.78±4.56ab | 314.21±15.37 | 140.56±12.03b | |
| g. 363T>C | TT | 69.68±5.47 | 76.16±4.83 | 301.38±12.82 | 145.73±14.54 |
| (MTNR1C) | TC | 70.43±5.53 | 76.53±4.82 | 303.74±14.12 | 145.68±15.67 |
| CC | 70.01±5.68 | 77.23±5.56 | 298.16±10.73 | 144.41±15.81 | |
Table 4
Association between polymorphisms in MTNR1A, MTNR1B, and MTNR1C genes and egg quality traits in Shaoxing ducks1)
REFERENCES
1. Kharwar RK, Haldar C. Anatomical and histological profile of bronchus-associated lymphoid tissue and localization of melatonin receptor types (Mel1a and Mel1b) in the lung-associated immune system of a tropical bird, Perdicula asiatica
. Acta Histochem 2011; 113:333–9.


2. Trivedi AK, Kumar V. Melatonin: an internal signal for daily and seasonal timing. Indian J Exp Biol 2014; 52:425–37.

3. Yadav SK, Haldar C. Reciprocal interaction between melatonin receptors (Mel1a, Mel1b, and Mel1c) and androgen receptor (AR) expression in immunoregulation of a seasonally breeding bird, Perdicula asiatica: role of photoperiod. J Photochem Photobiol B Biol 2013; 122:52–60.

4. Reppert SM. Melatonin receptors: molecular biology of a new family of G protein-coupled receptors. J Biol Rhythm 1997; 12:528–31.

5. Li DY, Smith DG, Hardeland R, et al. Melatonin receptor genes in vertebrates. Int J Mol Sci 2013; 14:11208–23.



6. Reppert SM, Weaver DR, Cassone VM, Godson C, Kolakowski LF. Melatonin receptors are for the birds: Molecular analysis of two receptor subtypes differentially expressed in chick brain. Neuron 1995; 15:1003–15.


7. Sundaresan NR, Marcus Leo MD, Subramani J, et al. Expression analysis of melatonin receptor subtypes in the ovary of domestic chicken. Vet Res Commun 2009; 33:49–56.


8. Wang Y, Peng K, Li J, et al. Ultrastructure and melatonin 1a receptor distribution in the ovaries of African ostrich chicks. Cytotechnology 2008; 56:187–95.



9. He H, Jiang DM, Kang B, et al. Gene expression profiling of melatonin receptor subtypes in the ovarian hierarchical follicles of the Sichuan white goose. Anim Reprod Sci 2014; 145:62–8.


10. Tsutsui K. A new key neurohormone controlling reproduction, gonadotropin-inhibitory hormone (GnIH): biosynthesis, mode of action and functional significance. Prog Neurobiol 2009; 88:76–88.


11. Ubuka T, Bentley GE, Ukena K, Wingfield JC, Tsutsui K. Melatonin induces the expression of gonadotropin-inhibitory hormone in the avian brain. Proc Natl Acad Sci USA 2005; 102:3052–7.



12. Chen H, Liu J, Fu Y. The study of Shaoxing duck. Chinese J Anim Sci 2003; 1:48–9.
13. Alsiddig MA, Yu SG, Pan ZX, et al. Association of single nucleotide polymorphism in melatonin receptor 1A gene with egg production traits in Yangzhou geese. Anim Genet 2017; 48:245–9.


14. Li DY, Zhang L, Smith DG, et al. Genetic effects of melatonin receptor genes on chicken reproductive traits. Czech J Anim Sci 2013; 58:58–64.


15. Niknafs S, Nejati-Javaremi A, Mehrabani-Yeganeh H, Fatemi SA. Estimation of genetic parameters for body weight and egg production traits in Mazandaran native chicken. Trop Anim Health Prod 2012; 44:1437–43.


16. Savegnago RP, Caetano SL, Ramos SB, et al. Estimates of genetic parameters, and cluster and principal components analyses of breeding values related to egg production traits in a White Leghorn population. Poult Sci 2011; 90:2174–88.



17. Goraga ZS, Nassar MK, Brockmann GA. Quantitative trait loci segregating in crosses between New Hampshire and White Leghorn chicken lines: I. egg production traits. Anim Genet 2012; 43:183–9.


18. Poivey JP. Heritabilities and genetic correlations of laying performance in Muscovy ducks selected in Taiwan. Br Poult Sci 2004; 45:180–5.


19. Cheng YS, Rouvier R, Poivey JP, Tai C. Genetic parameters of body weight, egg production and shell quality traits in the Brown Tsaiya laying duck. Genet Sel Evol 1995; 27:1–14.

20. Lin RL, Chen HP, Rouvier R, Marie-Etancelin C. Genetic parameters of body weight, egg production, and shell quality traits in the Shan Ma laying duck (Anas platyrhynchos). 2016; 95:2514–9.


21. El HM, Kang SW, Leclerc B, Kosonsiriluk S, Chaiseha Y. Dopamine-melatonin neurons in the avian hypothalamus and their role as photoperiodic clocks. Gen Comp Endocrinol 2009; 163:123–7.


22. Surbhi , Kumari Y, Rani S, Tsutsui K, Kumar V. Duration of melatonin regulates seasonal plasticity in subtropical Indian weaver bird, Ploceus philippinus
. Gen Comp Endocrinol 2015; 220:46–54.


23. Chowdhury VS, Yamamoto K, Ubuka T, et al. Melatonin stimulates the release of gonadotropin-inhibitory hormone by the avian hypothalamus. Endocrinology 2010; 151:271–80.



24. Rozenboim I, Aharony T, Yahav S. The effect of melatonin administration on circulating plasma luteinizing hormone concentration in castrated White Leghorn roosters. Poult Sci 2002; 81:1354–9.



25. Xu H, Zeng H, Luo C, et al. Genetic effects of polymorphisms in candidate genes and the QTL region on chicken age at first egg. BMC Genet 2011; 12:33



26. Feng XP, Kuhnlein U, Aggrey SE, Gavora JS, Zadworny D. Trait association of genetic markers in the growth hormone and the growth hormone receptor gene in a White Leghorn strain. Poult Sci 1997; 76:1770–5.



27. Dunn IC, Miao YW, Morris A, et al. A study of association between genetic markers in candidate genes and reproductive traits in one generation of a commercial broiler breeder hen population. Heredity 2004; 92:128–34.



28. Li HF, Shu JT, Du YF, et al. Analysis of the genetic effects of prolactin gene polymorphisms on chicken egg production. Mol Biol Rep 2013; 40:289–94.


29. Xu J, Gao X, Li X, et al. Molecular characterization, expression profile of the FSHR gene and its association with egg production traits in muscovy duck. J Genet 2017; 96:341–51.









 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print





