 |
 |
| Anim Biosci > Volume 31(2); 2018 > Article |
|
Abstract
Objective
The aim of this study was to investigate the effect of single nucleotide polymorphisms (SNPs) of the melanogenesis associated transcription factor (MITF) and dopachrome tautomerase (DCT) genes on plumage coloration in Asian native duck breeds. MITF encodes a protein for microphthalmia-associated transcription factor, which regulates the development and function of melanocytes for pigmentation of skin, hair, and eyes. Among the tyrosinase-related family genes, DCT is a pigment cell-specific gene that plays important roles in the melanin synthesis pathway and the expression of skin, feather, and retina color.
Methods
Five Asian duck varieties (black Korean native, white Korean native, commercial Peking, Nageswari, and Bangladeshi Deshi white ducks) were investigated to examine the polymorphisms associated with plumage colors. Among previously identified SNPs, three synonymous SNPs and one indel of MITF and nine SNPs in exon regions of DCT were genotyped. The allele frequencies for SNPs of the black and white plumage color populations were estimated and Fisher’s exact test was conducted to assess the association between the allele frequencies of these two populations.
Results
Two synonymous SNPs (c.114T>G and c.147T>C) and a 14-bp indel (GCTGCAAAC AGATG) in intron 7 of MITF were significantly associated with the black- and white-colored breeds (p<0.001). One non-synonymous SNP [c.938A>G (p.His313Arg)] in DCT, was highly significantly associated (p<0.001) and a synonymous SNP (c.753A>G) was significantly associated (p<0.05) with black and white color plumage in the studied duck populations.
The MITF gene encodes a protein known as the microphthalmia-associated transcription factor. The basic structure of MITF, which is located at the mi locus, consists of nine exons and a basic helix-loop-helix-leucine zipper (bHLH-LZ) protein structure [1]. The protein is involved in the development and function of melanocytes and is responsible for skin, hair, and eye color. This transcription factor facilitates the development of retinal pigment epithelial cells that contribute to retina color. Additionally, it also plays a role in the development of osteoclasts and mast cells in the bone marrow and regulates bone resorption and allergic reactions [1]. MITF consists of multiple isoforms with different amino termini derived from unique exon 1 sequences produced by alternative splicing [2]. In ducks, two types of MITF isoform (MITF-B and MITF-M), which are expressed in feather bulbs, have been reported [3].
MITF acts as a key regulator of melanogenesis enzyme genes of the Tyr family and melanocyte differentiation. Hyperpigmentation, as a natural mutant, was characterized in a Chinese native duck breed, Silky fowl [4]. In contrast, MITF may regulate hypopigmentation as well as patterning (white spotting) in both cattle and dog breeds [5,6]. In Japanese quail, the silver plumage color has been shown to be associated with the causal B mutation of MITF [7]. This mutation, resulting from a 2-bp deletion, produces a premature stop codon in exon 11 of MITF and is also associated with growth.
Among the genes of the tyrosinase-related protein ( TYRP) family, dopachrome tautomerase (DCT) plays important roles in the melanin pigmentation of vertebrates [8]. The TYRP2 protein was identified as the enzyme DCT at the slaty locus on chromosome 14 in mouse [9] and is known as either DCT or TYRP2 [10]. Expression of the DCT gene has been detected in the retina of ducks, whereas no expression was observed in black and white feather bulbs [3]. Hair melanin content was detected at a higher level in DCT knockout mice than in slaty mutant mice [11].
The National Institute of Animal Science in Korea has been conducting a conservation program to examine the genetic differences among indigenous duck breeds and has expanded selective breeding programs to improve performances with a view toward commercial production [12]. Likewise, a conservation scheme for the Bangladeshi indigenous duck breeds has also conducted for breed characterization and genetic improvement. In terms of industrial meat processing, white feather color has advantage over black feather color, in that the processed meat pigmentation and pin feathers are tend to be less preferred by consumers. Therefore, it is worthwhile to develop ducks with white feather color for economic advantage at the industrial level. In our previous study, we identified polymorphisms in different plumage color genes within Korean native black ducks and commercial meat-type Peking ducks [13].
Although the effects of MITF and DCT genes and their variations have previously been explored in humans, mice, and various other vertebrates, there have been few studies on these genes in livestock and poultry species, including ducks. Therefore, as a continuation of our research on the MITF and DCT genes, the present study was conducted to investigate polymorphisms in these two genes to assess their associations with different black and white plumage color in Asian duck breeds.
Five Asian duck breeds were selected for this study based on black and white plumage color. A total of 315 and 185 DNA samples were investigated from those five populations for DCT and MITF genes, respectively. Korean native duck blood samples were collected from the National Institute of Animal Science and the Chungnam National University farm in Korea. Blood samples of the two Bangladeshi duck varieties were collected from two governmental institutions as well as from duck farmers. Blood samples collected from Bangladeshi ducks were initially preserved on Whatman FTA classic cards (Whatman International Ltd, Maidstone, Kent, UK) and then the genomic DNA was isolated using a PrimePrep Genomic DNA Isolation Tissue Kit (GeNetBio, Daejeon, Korea) following the provided instructions. Genomic DNA of the Korean native and commercial Peking ducks was extracted from blood samples using a DNA Isolation Kit for blood (GeNetBio, Deajeon, Korea). The concentrations of extracted DNA were measured spectrophotometrically using a Thermo Scientific NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Stock DNA samples at a concentration of approximately 25 ng/μL were stored at −20°C for molecular analysis.
SNPs of the DCT and MITF genes and the primer sets used for polymerase chain reaction (PCR) amplification of the targeted SNPs were described in our previous study [13]. PCR-restriction fragment length polymorphism (PCR-RFLP) methods were used for genotyping the SNPs of the coding regions. The enzymatic reactions were performed in 20-μL reaction mixtures that included 15 μL of PCR products, 2 units of each restriction enzyme (Table 1), and reaction buffer. The digested products were separated electrophoretically on 3% agarose gels using 1×tris-acetate-ethylenediaminetetraacetic acid buffer and visualized with ethidium bromide. The KASP (LGC Tech, Teddington, Middlesex, UK) assay was designed for genotyping of the c.726C>T SNP of DCT. The allele discrimination PCR assay was based on an allele-specific primer pair using fluorescent dyes (FAM for the C allele and HEX for the T allele) and performed in an ABI 7500 Real Time PCR device (Applied Biosystems, Foster City, CA, USA) by following the KASP genotyping reaction guidelines.
The SNPs were filtered statistically with a minor allele frequency <5% and the allele frequencies of each SNP were examined to evaluate the associations with plumage colors using Fisher’s exact test by using PLINK software version 1.07 [14] at the p<0.05 level of significance. Additionally, Haploview software [15] was used to calculate haplotype numbers and their frequencies among seven SNPs from the four duck populations. In order to examine the association of haplotypes between two plumage color phenotypes, Fisher’s exact test was carried out using PHASE and R (version 3.3.0) software packages. The level of significance was set at p<0.05 for haplotype association.
Of the 36 SNPs of MITF identified in our previous study [13], only three synonymous SNPs located in exon 1 and 3 and one indel, which harbor restriction sites, were considered for SNP genotyping in the present study. Among these, two SNPs of MITF, c.147C>T and c.501A>G, were amplified for genotyping using primer sets used in our previous study [13]. Detailed information on the primers, SNP locations, and respective endonucleases of c.114T>G and g.42318-42331> GCTGCAAACAGATG are shown in Table 1. Two further exonic SNPs of MITF, c.117C>T and c.457C>T, were not considered for analysis in the present study due to the lack of restriction sites. For the c.114T>G polymorphism, digestion of an 811-bp DNA fragment with BstAPI enzyme produced fragments of 516, 217, and 78 bp for the G allele and fragments of 516 and 295 bp for the T allele (Figure 1A). For the c.147C>T SNP, the PCR products were digested with TspRI enzyme and cleaved into 569- and 11-bp fragments for the C allele and 402-, 167-, and 11-bp fragments for the T allele [13]. The c.501A>G polymorphism showed only a 424-bp fragment for the A allele and 285- and 139-bp fragments when digested with the NciI restriction enzyme (Figure 1B). A 14-bp, indel of MITF, g.42318-42331> GCTGCAAACAGATG, was identified using the restriction endonuclease Fnu4HI and the digestion of the PCR product resulted in fragments of 207, 94, and 63 bp for the I allele due to insertion, and 207 and 143 bp for the D allele (Figure 1C).
The genotype and allele frequencies of the identified indel and SNPs in exons of MITF from four duck populations categorized into two groups based on plumage color (white and black or mixed) are shown in Table 3. The black-colored duck group contained the Korean native black and Nageswari ducks, whereas Korean native white and commercial Peking ducks were grouped as the white-colored duck population. Detailed information on the plumage color and pattern of the four Asian duck breeds examined in the current study were presented in our previous study of MC1R [16]. For the c.114T>G polymorphism of MITF, the T allele frequency (0.352) was considerably higher in the black-colored duck population than in the white-colored ducks (0.144). In contrast, the white-colored duck population tended to have a higher G allele frequency (0.856) than the black-colored ducks (0.648) for this polymorphism, and the corresponding genotype frequencies in the white duck population were 0.476 (GG), 0.208 (GT), and 0.079 (TT). Similarly, the frequency of allele T for MITF c.147T>C SNP was predominantly higher in the black-colored duck population than in the white-colored ducks, whereas the C allele frequency in white-colored duck population was higher than that in the black-colored ducks. Moreover, the frequency of the heterozygous genotype (CT) for the c.147T>C SNP was higher than the homozygous genotype (CC) in the black-colored duck population and the TT genotype was completely absent for this mutation. In terms of the genotype and allele frequencies of the c.501A>G SNP of MITF located in exon 3, there were insignificant differences between the two duck populations, and the AA genotype was absent in the black-colored duck population. For the 14-bp indel (g.42318-42331> GCTGCAAACAG ATG), the allele frequency of D (deletion) in white-colored ducks was markedly higher than that in black-colored ducks and the DD genotype was not found in the Nageswari duck population (Table 2). Therefore, the frequency of allele I (insertion GCTGC AAACAGATG) was higher in the black-colored ducks than in the white-colored ducks and genotype frequencies of 0.845 (II), 0.119 (DI), and 0.036 (DD) were observed in the black-colored duck population. Collectively, the data for the genotyped ducks in the two groups indicate that the T allele was dominant to the G and C alleles for c.114T>G and c.147T>C SNPs, respectively.
In our previous investigation [13], we detected 30 polymorphisms by comparing the NCBI sequence of DCT (NCBI Gene ID: 10179 2699) with sequences obtained from the 5′ to 3′-untranslated region using seven primer sets [13]. Of these, 11 polymorphisms were identified from exons 1, 2, 4, and 5, seven of which were A↔G substitutions and four were C↔T transition substitutions. Furthermore, four substitutions were missense, whereas the others were synonymous [13]. Two SNPs of DCT, namely c.735C>T and c.935A>G, located in exon 4 and 5, respectively, have no restriction sites for target region genotyping. Therefore, nine SNPs of the coding region were genotyped in five Asian duck breeds by PCR-RFLP and KASP genotyping assay for further association study. The primer sets used for DCT gene amplification and the restriction enzymes used in RFLP are shown in Table 1. Three mutations in DCT at position c.468A>G, c.762C>T, and c.905C>T in exon 2, 4, and 5 respectively, were genotyped using restriction endonucleases. The NciI restriction patterns for c.468A>G were 294- and 179-bp fragments for the G allele and a single 473-bp fragment for the A allele. Genotyping of the c.762C>T polymorphism in exon 4 using the HpyCH4IV restriction site yielded a single 414-bp fragment for the T allele and 258- and 156-bp fragments for the C allele. Using the BsrDI restriction enzyme, the T allele for the c.905C>T polymorphism showed 416- and 84-bp fragments, whereas the C allele was represented by a single 500-bp fragment. The resultant RFLP fragments in the restriction profiles obtained for five other SNPs are shown in Figure 1 (D, E, F, G, and H). In addition, the PROVEN (http://provean.jcvi.org/seq_submit.php) program was used to investigate the effects on protein functions as a consequence of amino acid changes. Three variants (T302M, D312G, and R313H) located in exon 5 may have deleterious effects on protein function, whereas the R21H variant has a neutral (neither harmful nor beneficial) effect according to the PROVEN scores [17]. The genotyped data for each SNP of DCT of the five duck breeds are presented in Table 4. Moreover, Table 5 presents the number of birds genotyped and allele frequencies in the two different plumage color populations. The black-colored duck population comprised black Korean native and Nageswari ducks (146 in number), whereas white Korean native, Peking, and Deshi white ducks were grouped as the white-colored duck population (169 in number). Allele frequencies were calculated for nine genotyped SNPs of DCT using the black- and white-colored duck populations with total number of 314 birds.
Association studies between the two duck populations with distinct plumage colors were performed using genotyping data of four polymorphisms in 185 ducks. Significant association was found between SNPs c.114T>G and c.147C>T and a 14-bp indel and plumage color variants. In contrast, the allele frequency results for the c.501A>G (p.Pro167Pro) SNP in exon 3 showed a non-significant association with plumage color (Table 3). The number of ducks having the GG genotype (0.71) for c.114T>G (p.Val38Val) SNP was higher in the white-colored duck population. Similarly, the CC genotype frequency for the c. 147C>T SNP (p.Ser49Ser) was higher (0.64) in the white duck population. Therefore, the results indicated that the two alleles G and C for the SNPs c.114T>G and 147C>T, respectively, had a significant association with white plumage color. In contrast, black plumage color was significantly associated with allele I for an indel (g.42318-42331> GCTGCAAACAGATG) and the alleles T and G for the c.114T>G and 147C>T polymorphisms, respectively.
The results of association analyses of the nine genotyped SNPs for the two plumage color duck populations examined in this study are presented in the Table 5. One SNP, c.938A>G, located in exon 5 was highly significantly associated (p<0.001) with plumage color (Table 5). The c.753A>G SNP was also significantly associated (p<0.05) with plumage color, whereas no significant associations were observed between the other seven SNPs and black or white plumage color. The A allele frequency (0.58) for the c.938A>G SNP was higher in the white-colored plumage group, whereas the G allele frequency was higher (0.55) in the black-colored duck group. Therefore, the association results shown in Table 5 indicate that allele A for the substitution p.His313Arg is associated with white plumage color, whereas the G allele is associated with black plumage color.
Haplotypes were reconstructed based on three polymorphisms and an indel and their frequencies are presented in Table 6. A total of nine haplotypes were detected from four Asian duck populations. Two haplotypes (GCGI and GTGI) were found in all duck breeds, whereas TTGI and GCAI haplotypes were only observed in Nageswari and white Korean native ducks, respectively. Haplotype GCGD had the highest frequency (60.1%) in commercial (Peking) ducks, low frequencies in white Korean native ducks (9.4%) and black Korean native ducks (4.2%), and was absent in Nageswari ducks. The haplotype GCGI was distributed across all duck populations, having the highest frequency in Nageswari ducks (55.6%), followed by frequencies of 24.9%, 22.6%, and 15.6% in white Korean native, Peking, and black Korean native ducks, respectively. The GTGI haplotype had a distribution pattern similar to that of the GCGI haplotype. Both white Korean native and black Korean native duck varieties contained eight haplotypes. The haplotypes TCGI, TCGD, and TCAI were restricted to the Korean native duck populations. Haploview analysis [15] using 4 SNPs of MITF genes showed high linkage disequilibrium (LD) among themselves (r2≥0.62) except between variants c.147T>C and g.42318~31(r2≥0.38). The moderate to high LD coefficient revealed potential non-random association among the SNPs of this gene (data not shown). The results of association analyses between plumage color phenotypes and haplotypes are shown in Table 7. The GCGD haplotype of MITF was significantly associated (p<0.005) with a white plumage color phenotype, whereas two haplotypes, TCGI and GTGI, have significant associations with a black- or mixed-colored plumage phenotype. The remaining four haplotypes showed no association with plumage color.
Although several previous studies have reported specific association of SNPs of MITF in cattle [18], buffalo [19], and dog [20], there is virtually no information on the SNPs of MITF in poultry, including duck. The present study, to the best of our knowledge, is the first to report an SNP-specific association analysis to discriminate between Asian ducks with black and white plumage color. Previously, a 2-bp deletion in exon 11 of the quail MITF gene was shown to generate a premature stop codon and resulted in a B mutation for silver plumage color [7]. In the current study, two synonymous SNPs in exon 1 and a 14-bp indel in intron 7 were found to have a highly significant association (p<0.001) with the plumage color of ducks. The intronic indel and SNPs located in exon 1 might be associated with plumage color variants or be closely related to other SNPs in the regulatory region of this gene and/or other adjacent SNPs in pigment cell-specific genes. Our results are supported by the findings of previous studies by Kimchi-Sarfaty et al [21], Komar [22], and Fernandez-Calero et al [23]. These authors reported that silent polymorphisms that did not alter amino acid sequences in the human multidrug resistance 1 and estrogen receptor alpha genes could change the substrate specificity of a protein because of different structural and functional properties of a substrate, which might have an impact on a particular phenotype.
In the present study, the average genotype frequency of the 14-bp insertion was greater (0.85) in the black-colored duck population than that observed in the white-colored duck population. An 18-bp insertion located between exon 5 and 6 was identified in the dog MITF-M+ isoform, which was associated with the addition of six amino acids in the polypeptide chain [20]. The MITF-M isoform activated melanocytes in the skin of the white Samoyed breed of dog. In many dog breeds, random white spotting has been attributed to an insertion of a short interspersed nucleotide at the 5′ end of the MITF-M gene [6,24]. Li et al [3], however, reported contrasting results in ducks. They reported that expression of both MITF-M and MITF-B isoforms was observed in the black feather bulbs and retinas of ducks, whereas the expression of MITF-M was not detected in white feather bulbs. In addition, their results showed that cis- or trans-acting regulatory elements affected the MITF-M isoform to determine the plumage pigmentation in ducks, which is in contrast to observations for the non-synonymous polymorphisms in Japanese quail. This conformational change in protein substrates might be associated with similar gene expression pattern in our studied duck population.
Although several studies have examined the gene expression and molecular character of MITF, there have been very few studies on SNP polymorphisms and their associations with pigmentation in livestock animals. A missense mutation (p. Arg210Ile) in the bovine MITF gene traced to chromosome 22 was shown to be the cause of German White Fleckvieh syndrome, which is dominantly inherited in Fleckvieh cattle and associated with hypopigmentation [18]. In a recent study on swamp buffalo, a nonsense mutation (p. Arg110*) in exon 3 and a donor splice-site mutation (p. Glu281_Leu282Ins8) in intron 7 of MITF were mapped to chromosome 22, both of which are associated with white-spotted coat color [19]. Similarly, in six Ethiopian cattle populations with different environmental adaptation, it was shown that coat color variation was associated with MITF, and a high genetic distance was observed between spotted and unspotted populations [24]. The A allele frequency for the rs137845005 SNP of MITF was observed at a higher percentage in spotted coat color populations, whereas the G allele frequency was greater in cattle populations with uniform coat color. These results are partially consistent with our findings regarding two SNPs (c.114T>G and c.147C>T) in exon 1, which were significantly associated with plumage color in white and black feather color duck populations.
In the MITF gene, the average homozygous genotypes (TT) for these two SNPs were higher in the mixed- or black-colored duck population. Moreover, the average heterozygous genotype frequency for the c.147C>T SNP was higher (0.63) in the black-colored duck population and lower (0.36) in the white-colored duck population. The constructed haplotype results of this study showed that the GCGD haplotype had the highest frequency in the white-colored duck population. The GCGI and TCGI haplotypes showed the second and third highest haplotype frequencies, respectively, which are associated with the black plumage color phenotype in Nageswari and black Korean native ducks, respectively. Notably, three haplotypes, TCGI, TCGD, and TCAI, were only observed in the Korean native black and white color duck breeds. Generally, reconstructed haplotypes provide more explicit results compared to a particular SNP found in a specific region of the gene or genome. For example, haplotypes accounted for 80% more phenotypic variance in adiponectin, C1Q and collagen domain containing (ADIPOQ), calpain 1 (CAPN1), C-X-C chemokine receptor type 4 (CXCR4), CCAAT/enhenhancer- binding protein alpha (CEBPA), and fatty acid synthase (FASN) genes and showed a robust association with intramuscular fat content in Bos taurus cattle compared to single SNP analyses [25], which is consistent with the findings of the present study. Such assembled haplotypes could be utilized for the selection and development of specific varieties with distinctive phenotypes.
Numerous studies have been conducted on the molecular characterization and gene expression of DCT associated with pigmentation in mice [8,26], chicken [27], quail [28], cattle [29], and duck [3]. We identified polymorphisms of this gene and their association with plumage color in different Asian duck breeds for the first time. Li et al [3] reported that DCT was expressed only in the retina, whereas all ducks used in the experiment had normal dark retinas. However, no expression of this gene was identified in the white and black feather bulbs of ducks with white, black, and white-black dorsal plumage. DCT consists of 8 exons and spans 17.3 kb of the duck genome; whereas its chromosomal location in duck has yet to be determined [30]. In the present study, one SNP, c.938A>G, in exon 5 was significantly associated (p>0.001) with black and white plumage color in ducks. The frequency of the G allele of this c.938A>G polymorphism was higher in ducks with black-colored plumage than in those with white-colored plumage, whereas the A allele frequency was higher in white ducks. These results indicate that the amino acid substitution p.His313Arg might be associated with plumage pigmentation. Similar results have been reported in previous studies of mice coat color. A single amino acid substitution (p. Arg194Gln) was identified in the first copper binding site of the TYRP2 protein of slaty mice [8]. This non-synonymous mutation was associated with a reduction in the relative activity of DCT by more than 3 = fold at the slaty locus compared with that in wild type mice due to the conversion of DOPAchrome to 5,6-dihydroxyindole carboxylic acid in melanin synthesis. One deletion and three polymorphisms in DCT of mice are responsible for the dark gray coat color [11]. Three mutations identified at the DCT locus in mice were shown to cause different pigmentation phenotypes [26]. The slaty-2J mutation, a point mutation in exon 7, was characterized as recessive to the wild type and results in the substitution of proline by leucine, whereas the slaty-light mutation results in the change of arginine to glycine at base 486 [26]. Guyonneau et al [11] reported restricted coat color effects due to deficient production of the DCT protein in knockout mice for this gene. However, in French cattle breeds, the expression patterns of TYR, TYRP1, and DCT genes showed no correlation with coat color dilution [29].
In conclusion, we identified SNPs in MITF and examined their association with black and white plumage color in duck varieties. Among these, the two alleles c.114G and c.147C in exon 1 and an indel, g.42318-42331> GCTGCAAACAGATG, may have the effect of enhancing MITF activity and, consequently, eumelanin deposition in duck plumage. Two significant novel mutations that we identified in the duck DCT gene, a c.752A>G synonymous mutation in exon 4 and the ce.938A>G SNP in exon 5, might have effects on plumage coloration as a consequence of the replacement of histidine with arginine, although further investigations using large populations are required to verify this. However, absence of plumage color specific alleles or haplotypes limit the scope for complete separation of black from white colored duck. Overall, these mutations could, therefore, be considered as potential markers for selection of desired colors in duck populations.
ACKNOWLEDGMENT
This work was supported by research fund of Chungnam National University (2015-1065-01).
Figure 1
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) genotyping patterns of two single nucleotide polymorphisms (SNPs) and one indel (A, B, and C) in the melanogenesis associated transcription factor (MITF) gene; and five SNPs in the coding region (D, E, F, G, and H) of dopachrome tautomerase (DCT). Arrows in the chromatograms indicate the polymorphic sites. M: 100-bp molecular size marker.
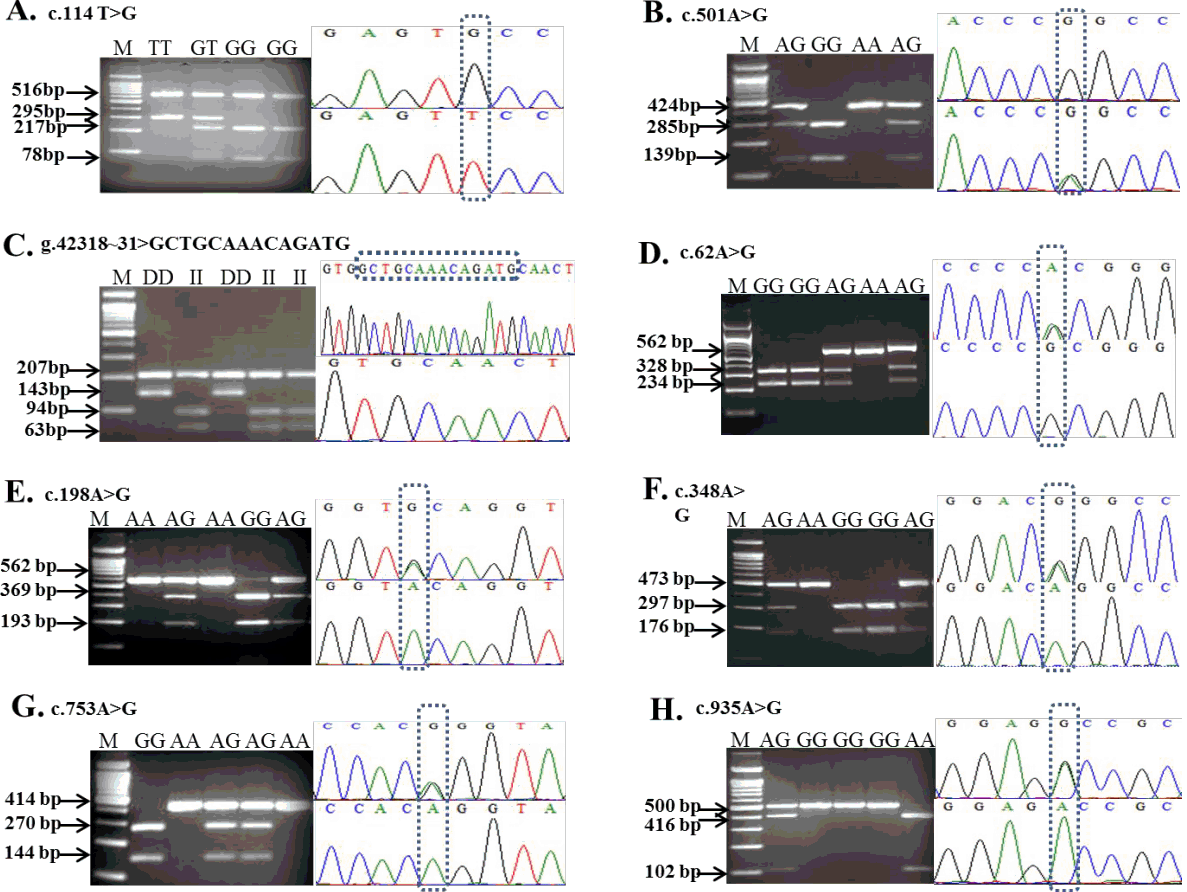
Table 1
Primer information and the restriction enzymes used in this study
Table 2
Genotype distribution of SNPs in the MITF gene of different colored duck breeds
Table 3
The results of association analysis of SNPs in the MITF gene between black and white duck breeds
| Population/group1) | No. of birds | SNP position | Genotype frequency | Allele frequency | p-value | |||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| GG | GT | TT | T | G | ||||
| Black-colored ducks | 84 | c.114T>G | 0.476 | 0.345 | 0.179 | 0.352 | 0.648 | 0.0016* |
| White-colored ducks | 101 | 0.713 | 0.208 | 0.079 | 0.144 | 0.856 | ||
|
|
|
|||||||
| CC | CT | TT | T | C | ||||
|
|
|
|||||||
| Black-colored ducks | 84 | c.147T>C | 0.369 | 0.631 | 0.000 | 0.315 | 0.685 | 0.0002* |
| White-colored ducks | 101 | 0.644 | 0.356 | 0.000 | 0.178 | 0.822 | ||
|
|
|
|||||||
| GG | AG | AA | A | G | ||||
|
|
|
|||||||
| Black-colored ducks | 84 | c.501A>G | 0.833 | 0.167 | 0.000 | 0.083 | 0.917 | 0.888 |
| White-colored ducks | 101 | 0.861 | 0.119 | 0.020 | 0.079 | 0.921 | ||
|
|
|
|||||||
| II | DI2) | DD | D | I | ||||
|
|
|
|||||||
| Black-colored ducks | 84 | g.42318~31 | 0.845 | 0.119 | 0.036 | 0.012 | 0.988 | 2.44e-08* |
| White-colored ducks | 101 | 0.406 | 0.238 | 0.356 | 0.476 | 0.524 | ||
Table 4
Genotype distribution of SNPs in DCT of different duck breeds
| Breed/varity1) | No. of birds | SNP position | Genotype (Number of birds) | ||
|---|---|---|---|---|---|
|
|
|||||
| GG | AG | AA | |||
| BKND | 110 | c.62A>G | 57 | 43 | 10 |
| WKND | 90 | 40 | 41 | 9 | |
| CD | 59 | 15 | 37 | 7 | |
| BaB | 36 | 11 | 14 | 11 | |
| BaW | 20 | 6 | 8 | 6 | |
|
|
|||||
| GG | AG | AA | |||
|
|
|||||
| BKND | 110 | c.198A>G | 10 | 44 | 56 |
| WKND | 90 | 9 | 40 | 41 | |
| CD | 59 | 7 | 36 | 16 | |
| BaB | 36 | 12 | 14 | 10 | |
| BaW | 20 | 3 | 8 | 9 | |
|
|
|||||
| GG | AG | AA | |||
|
|
|||||
| BKND | 110 | c.348A>G | 28 | 63 | 19 |
| WKND | 90 | 28 | 44 | 18 | |
| CD | 59 | 5 | 29 | 25 | |
| BaB | 36 | 6 | 14 | 16 | |
| BaW | 20 | 9 | 8 | 3 | |
|
|
|||||
| GG | AG | AA | |||
|
|
|||||
| BKND | 110 | c.468A>G | 46 | 62 | 2 |
| WKND | 90 | 70 | 18 | 2 | |
| CD | 59 | 34 | 21 | 4 | |
| BaB | 36 | 28 | 6 | 2 | |
| BaW | 20 | 20 | 0 | 0 | |
|
|
|||||
| CC | CT | TT | |||
|
|
|||||
| BKND | 110 | c.726C>T | 42 | 56 | 12 |
| WKND | 90 | 37 | 42 | 11 | |
| CD | 59 | 13 | 20 | 26 | |
| BaB | 36 | 14 | 15 | 7 | |
| BaW | 20 | 10 | 10 | 0 | |
|
|
|||||
| GG | AG | AA | |||
|
|
|||||
| BKND | 110 | c.753A>G | 49 | 49 | 12 |
| WKND | 90 | 33 | 45 | 12 | |
| CD | 59 | 15 | 23 | 21 | |
| BaB | 36 | 14 | 15 | 7 | |
| BaW | 20 | 10 | 10 | 0 | |
|
|
|||||
| CC | CT | TT | |||
|
|
|||||
| BKND | 110 | c.762C>T | 10 | 49 | 51 |
| WKND | 90 | 12 | 42 | 36 | |
| CD | 59 | 10 | 35 | 14 | |
| BaB | 36 | 7 | 15 | 14 | |
| BaW | 20 | 0 | 10 | 10 | |
|
|
|||||
| CC | CT | TT | |||
|
|
|||||
| BKND | 110 | c.905C>T | 39 | 49 | 22 |
| WKND | 90 | 11 | 50 | 29 | |
| CD | 59 | 11 | 32 | 16 | |
| BaB | 36 | 2 | 9 | 25 | |
| BaW | 20 | 2 | 8 | 10 | |
|
|
|||||
| GG | AG | AA | |||
|
|
|||||
| BKND | 110 | c.938A>G | 44 | 50 | 16 |
| WKND | 90 | 11 | 51 | 28 | |
| CD | 59 | 11 | 32 | 16 | |
| BaB | 36 | 5 | 12 | 19 | |
| BaW | 20 | 3 | 8 | 9 | |
Table 5
Genotype distribution of SNPs in DCT between black- and white-colored duck breeds
| Group/population1) | No. of bird | SNP position | Genotype (Number of bird) | Allele frequency | p-value | |||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| GG | AG | AA | G | A | ||||
| Black color duck | 146 | c.62A>G | 68 | 57 | 21 | 0.66 | 0.34 | 0.246 |
| White color duck | 169 | 61 | 86 | 22 | 0.62 | 0.38 | ||
|
|
|
|||||||
| GG | AG | AA | G | A | ||||
|
|
|
|||||||
| Black color duck | 146 | c.198A>G | 22 | 58 | 66 | 0.35 | 0.65 | 0.802 |
| White color duck | 169 | 19 | 84 | 66 | 0.36 | 0.64 | ||
|
|
|
|||||||
| GG | AG | AA | G | A | ||||
|
|
|
|||||||
| Black color duck | 146 | c.348A>G | 34 | 77 | 35 | 0.50 | 0.50 | 0.873 |
| White color duck | 169 | 42 | 81 | 46 | 0.49 | 0.51 | ||
|
|
|
|||||||
| GG | AG | AA | G | A | ||||
|
|
|
|||||||
| Black color duck | 146 | c.468A>G | 74 | 68 | 4 | 0.74 | 0.26 | 0.449 |
| White color duck | 169 | 124 | 39 | 6 | 0.85 | 0.15 | ||
|
|
|
|||||||
| CC | CT | TT | C | T | ||||
|
|
|
|||||||
| Black color duck | 146 | c.726C>T | 56 | 71 | 19 | 0.63 | 0.37 | 0.143 |
| White color duck | 169 | 60 | 72 | 37 | 0.66 | 0.34 | ||
|
|
|
|||||||
| GG | AG | AA | G | A | ||||
|
|
|
|||||||
| Black color duck | 146 | c.753A>G | 63 | 64 | 19 | 0.65 | 0.35 | 0.049* |
| White color duck | 169 | 58 | 78 | 33 | 0.66 | 0.34 | ||
|
|
|
|||||||
| CC | CT | TT | C | T | ||||
|
|
|
|||||||
| Black color duck | 146 | c.762C>T | 17 | 64 | 65 | 0.34 | 0.66 | 0.185 |
| White color duck | 169 | 22 | 87 | 60 | 0.39 | 0.61 | ||
|
|
|
|||||||
| CC | CT | TT | C | T | ||||
|
|
|
|||||||
| Black color duck | 146 | c.905C>T | 41 | 58 | 47 | 0.48 | 0.52 | 0.077 |
| White color duck | 169 | 24 | 90 | 55 | 0.41 | 0.59 | ||
|
|
|
|||||||
| GG | AG | AA | G | A | ||||
|
|
|
|||||||
| Black color duck | 146 | c.938A>G | 49 | 62 | 35 | 0.55 | 0.45 | 0.001** |
| White color duck | 169 | 25 | 91 | 53 | 0.42 | 0.58 | ||
Table 6
Reconstructed haplotypes and their frequencies for the MITF gene of four duck breeds
| Haplotype | Haplotype frequencies1) (%) | |||
|---|---|---|---|---|
|
|
||||
| BKND | BaB | CD | WKND | |
| GCGD | 4.2 | - | 60.1 | 9.4 |
| GCGI | 15.6 | 55.6 | 22.6 | 24.9 |
| TCGI | 41.9 | - | - | 21.9 |
| GTGD | 3.0 | 9.7 | 10.5 | - |
| GTGI | 18.6 | 30.6 | 6.7 | 18.7 |
| TCAI | 14.4 | - | - | 16.1 |
| TCGD | 2.1 | - | - | 6.1 |
| TTGI | - | 4.2 | - | - |
| GCAI | - | - | - | 2.6 |
Table 7
The results of the haplotype associations in the MITF locus between black and white color phenotypes
| Haplotype | Number of bird | p-value | |
|---|---|---|---|
|
|
|||
| Black or mixed plumage color | White plumage color | ||
| GCGD | 11 | 81 | 9.945e-09* |
| GCGI | 45 | 46 | 0.6079 |
| TCGI | 43 | 18 | 0.0006* |
| GTGD | 3 | 10 | 0.1523 |
| GTGI | 50 | 26 | 0.0040* |
| TCAI | 14 | 14 | 0.6877 |
| TCGD | 2 | 5 | 0.4627 |
REFERENCES
1. Hodgkinson CA, Moore KJ, Nakayama A, et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic helix-loop-helix-zipper protein. Cell 1993; 74:395–404.


2. Fuse N, Yasumoto K, Takeda K, et al. Molecular cloning of cDNA encoding a novel microphthalmia-associated transcription factor isoform with a distinct amino terminus. J Biochem 1999; 126:1043–51.



3. Li S, Wang C, Yu W, Zhao S, Gong Y. Identification of genes related to white and black plumage formation by RNA-seq from white and black feather bulbs in ducks. PLoS ONE 2012; 7:e36592



4. Li Y, Zhu X, Yang L, et al. Deng Expression and network analysis of genes related to melanocyte development in the Silky Fowl and White Leghorn embryos. Mol Biol Rep 2011; 38:1433–41.


5. Fontanesi L, Scotti E, Russo V. Haplotype variability in the bovine MITF gene and association with piebaldism in Holstein and Simmental cattle breeds. Anim Genet 2012; 43:250–6.


6. Karlsson EK, Baranowska I, Wade CM, Salmon HN, Zody MC. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet 2007; 39:1321–8.


7. Minvielle F, Bed’Hom B, Coville JL, et al. The “silver” Japanese quail and the MITF gene: causal mutation, associated traits and homology with the “blue” chicken plumage. BMC Genet 2010; 11:15



8. Jackson I, Chambers D, Tsukamoto K, et al. A second tyrosinase-related protein, TRP-2, maps to and is located at the mouse slaty locus. EMBO J 1992; 11:527–35.



9. Kroumpouzos G, Urabe K, Kobayashi T, Sakai C, Hearing VJ. Functional analysis of the slaty gene product (TRP2) as dopachrome tautomerase and the effect of a point mutation on its catalytic function. Biochem Biophys Res Commun 1994; 202:1060–8.


10. Tsukamoto K, Jackson IJ, Urabe K, Montague PM, Hearing VJ. A second tyrosinase-related protein, TRP-2, is a melanogenic enzyme termed DOPAchrome tautomerase. EMBO J 1992; 11:519–26.



11. Guyonneau L, Murisier F, Rossier A, Moulin A, Beermann A. Melanocytes and pigmentation are affected in dopachrome tautomerase knockout mice. Mol Cell Biol 2004; 24:3396–403.



12. Kim HR, Kwon HJ, Oh ST, et al. Effect of dietary metabolizable energy and crude protein concentrations on growth performance and carcass characteristics of Korean native ducks. Korean J Poult Sci 2012; 39:167–75.


13. Sultana H, Seo DW, Park HB, et al. Identification of polymorphisms in plumage color related genes in Korean native duck. J Fac Agr Kyushu U 2015; 60:119–26.
14. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 2007; 81:559–75.



15. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21:263–5.



16. Sultana H, Seo DW, Park HB, et al. Identification of MC1R SNPs and their association with plumage colors in Asian duck. J Poult Sci 2017; 54:111–20.



17. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012; 7:e46688



18. Philipp U, Lupp B, Momke S, et al. A MITF mutation associated with a dominant white phenotype and bilateral deafness in German Fleckvieh cattle. PLoS One 2011; 6:e28857



19. Yusnizar Y, Wilbe M, Herlino AO, et al. Microphthalmia-associated transcription factor mutations are associated with white-spotted coat color in swamp buffalo. Anim Genet 2015; 46:676–82.



20. Schmutz SM, Berryere TG, Dreger DL. MITF and white spotting in dogs: a population study. J Hered 2009; 100:66–74.



21. Kimchi-Sarfaty C, Oh JM, Kim IW, et al. A “Silent” polymorphism in the MDR1 gene changes substrate specificity. Science 2007; 315:5811525–8.


23. Fernandez-Calero T, Cabrera-Cabrera F, Ehrlich R, Marin M. Silent polymorphisms: can the tRNA population explain changes in protein properties? Life (Basel) 2016; 6:9



24. Edea Z, Kim KS.
MITF gene locus is associated with coat color variation of Ethiopian cattle populations adapted to different altitude environments. In : Proceedings, 10th World Congress of Genetics Applied to Livestock Production; 2014; Canada.
25. Barendse W. Haplotype analysis improved evidence for candidate genes for intramuscular fat percentage from a genome wide association study of cattle. PLoS ONE 2011; 6:e29601



26. Budd PS, Jackson IJ. Structure of the mouse tyrosinaserelated protein 2/dopachrome tautomerase (Tyrp2/Dct) gene and sequence of two novel slaty alleles. Genomics 1995; 29:35–43.


27. April CS, Jackson IJ, Kidson SH. Molecular cloning and sequence analysis of a chicken cDNA encoding tyrosinase-related protein-2/DOPAchrome tautomerase. Gene 1998; 219:45–53.


28. Niwa T, Mochii M, Nakamura A, Shiojiri N. Plumage pigmentation and expression of its regulatory genes during quail development- histochemical analysis using Bh (black at hatch) mutants. Mech Dev 2002; 118:139–46.








 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print





