 |
 |
| Anim Biosci > Volume 30(2); 2017 > Article |
|
Abstract
Objective
This study was designed to characterize the DNA polymorphisms of the melanocortin-1 receptor (MC1R) gene in indigenous Saudi Arabian sheep breeds exhibiting different color coats, along with individuals of the Sawaknee breed, an exotic sheep imported from Sudan.
Methods
The complete coding region of MC1R gene including parts of 3′ and 5′ untranslated regions was amplified and sequenced from three the indigenous Saudi sheep; Najdi (generally black, n = 41), Naeimi (generally white with brown faces, n = 36) and Herri (generally white, n = 18), in addition to 13 Sawaknee sheep.
Results
Five single nucleotide polymorphisms (SNPs) were detected in the MC1R gene: two led to nonsynonymous mutations (c.218 T>A, p.73 Met>Lys and c.361 G>A, p.121 Asp>Asn) and three led to synonymous mutations (c.429 C>T, p.143 Tyr>Tyr; c.600 T>G, p.200 Leu>Leu, and c.735 C>T, p.245 Ile>Ile). Based on these five SNPs, eight haplotypes representing MC1R Ed and E+ alleles were identified among the studied sheep breeds. The most common haplotype (H3) of the dominant Ed allele was associated with either black or brown coat color in Najdi and Sawaknee sheep, respectively. Two other haplotypes (H6 and H7) of Ed allele, with only the nonsynonymous mutation A218T, were detected for the first time in Saudi indigenous sheep.
Genetic variability assessment within and among different sheep breeds is essential to develop a successful breeding program for selecting ewes that express superior production traits, e.g. those concerning meat, milk and wool production [1]. Saudi Arabia has three native sheep breeds, namely Najdi, Naeimi, and Herri. The Najdi and Naeimi sheep are fat-tailed, adaptable to prevailing adverse environments and are considered the breeds of choice among Saudi consumers [2]. The Herri sheep breed that has also noticeable adaptability to prevailing adverse environments is an excellent indigenous breed in Saudi Arabia. The genetic determinants of coat color in farm animals, including sheep, are critical for breed recognition and fiber production. Coat color in farm animals is divergent and bears significant biological and economic impacts [3,4]. Melanin is the substance produced by melanocytes that gives skin, hair, and eyes their color. In a large number of mammalian species, the coat color diversity is mainly determined by the relative amount of the two basic melanins, eumelanin (black/brown) and pheomelanin (yellow/red), which are genetically controlled by the extension (E) and agouti (A) loci, respectively [5]. The A locus encodes for the agouti signaling protein (ASIP), a small paracrine signaling molecule [6]. The E locus encodes for the melanocortin-1 receptor (MC1R), a transmembrane protein with seven domains belonging to the G protein-coupled membrane receptors, and localizes on the surface of the melanocyte membrane [7]. When MC1R receptor is activated, it triggers a series of chemical reactions inside melanocytes that stimulate these cells to make eumelanin. If the receptor is not activated or blocked, melanocytes make pheomelanin instead of eumelanin. It has been found that the DNA polymorphisms in MC1R gene may reduce the ability of MC1R to stimulate eumelanin production, causing melanocytes to make mostly pheomelanin [8].
In sheep, the MC1R gene is located on chromosome 14 and has three main alleles, namely E+, Ed, and e, which are defined by three mutations in the coding region and associated with variation in coat color [9–12]. Studies of MC1R gene have provided valuable insights not only into the biology of pigmentation but also the evolution of domesticated animals [13]. So far, there is no report regarding the potential association of MC1R mutations with coat colors in Saudi indigenous sheep. Therefore, the objective of this study was to characterize the DNA polymorphisms of MC1R gene and assess their effects on Saudi Arabian sheep breeds exhibiting different color coats. Individuals of Sawaknee sheep, exotic breed imported from Sudan, were included in this study.
To assess MC1R genetic diversity in Saudi sheep, a total of 108 animals were selected from eleven flocks representing three indigenous breeds, namely Najdi (n = 41), Naeimi (n = 36), and Herri (n = 18), in addition to 13 Sawaknee sheep, originally from Sudan. Ten mL of blood samples were collected from each individual sheep by jugular venipuncture into vacuum EDTA tubes. Genomic DNA was extracted using the QIAgen DNeasy blood and tissue DNA extraction kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The quantity and quality of DNA were checked by Jenway Genova spectrophotometer (Krackeler Scientific Incorporated, Albany, NY, USA). The O.D. 260/280 ratios were between 1.7 and 1.9 indicating high quality DNA [14].
The two primers MF (5′-GAGAGCAAGCACCCTTTCCT-3′) and MR (5′-GAGAGTCCTGTGATTCCCCT-3′) were used to amplify the complete coding region of MC1R gene from the 108 individual sheep [15]. Polymerase chain reaction (PCR) amplifications were carried out in a 25-μL reaction volume containing 100 ng of template DNA and 20 pmol of each primer. To reduce the possibility of cross contamination and variation in the amplification reactions, master mixes containing all PCR reagents including the Kapa Taq polymerase enzyme (KAPA Biosystems, Boston, MA, USA) except DNA template were used. The amplification program was performed using the Gene Amp 9700 thermocycler (Applied Biosystems, Warrington, UK). The amplification protocol was an initial denaturation step for 2 min at 94°C, followed by 35 cycles of 94°C denaturation step for 0.5 min, 60°C annealing step for 0.6 min and 72°C extension step for 1 min. The final step was an extension step at 72°C for 5 min. Electrophoresis of the PCR products was done using 1.5% agarose gel and bands were detected by UV lamp after ethidium bromide staining using gel documentation system (Amersham Biosciences, Uppsala, Sweden).
PCR products of MC1R fragments were cleaned and sequenced by the Advanced Genetic Technologies Center (http://www.uky.edu/Centers/AGTC/). The DNA sequences were edited and aligned using BioEdit software (http://www.mbio.ncsu.edu/Bioedit/bioedit.html). The Basic Local Alignment Search Tool (BLAST) was used to search the GenBank database (http:www.ncbi.nlm.nih.gov) for homologous sequences. The BioEdit software was also used to detect single nucleotide polymorphisms (SNPs) and mutations. MC1R haplotypes and genotypes were determined within and among the studied sheep breeds using BioEdit software.
The amplification of the MC1R gene generated a PCR product of 1,170 bp in length from 41 Najdi, 36 Naeimi, 18 Herri, and 13 Sawaknee sheep. After sequencing, cleaning and aligning the MC1R region from the 108 animals, no SNPs were detected in either the 5′ or 3′ untranslated regions. However, five SNPs were detected in the MC1R complete open reading frame (ORF; 954 bp). Two SNPs were nonsynonymous mutations (c. A218T, p. Met73Lys and c. A361G, p. Asp121Asn), whereas the other three SNPs were synonymous (c. C429T, p. Tyr143Tyr; c. T600G, p. Leu200Leu. and c. C735T, p. Ile245Ile).
Each of the five SNPs had two alleles with three different genotypes (Tables 1 and 2). Generally, the allele frequencies of the five positions differed among the studied sheep breeds. Although, all possible alleles of the three synonymous SNPs were detected in the three indigenous Saudi sheep breeds, the Sawaknee breed, imported from Sudan, had only one allele at the five positions (Table 1). In Najdi sheep, the most common allele at position C429T was “T” with a frequency of 91% followed by 0.61% in Herri, while the frequency of the other allele “C” was 0.83% in Naeimi sheep (Table 1). Same pattern was seen in the other two synonymous SNPs (Table 1). The nonsynonymous SNPs, associated with dominant black coat color, at 218 and 361 positions were found in higher frequencies (0.83 and 0.75, respectively) in Najdi breed (generally black with white faces) (Table 1; Figure 1). These two alleles were fixed in Sawaknee sheep (Table 1). In contrast, the Naeimi breed (generally white with brown faces) lacked the A allele at 361 position, where the G allele was fixed (Table 1; Figure 1). The genotype frequencies of the five SNPs varied also among the three Saudi breeds. No heterozygous genotypes of the five SNPs were detected in Sawaknee sheep (generally brown) (Table 2; Figure 1). However, heterozygous genotypes at the five SNP positions were detected in the indigenous Saudi sheep (Table 2). Generally, Najdi sheep showed the highest allelic variation in the MC1R gene, followed by Herri then Naeimi. Sawaknee sheep showed the lowest allelic variation where five out of the 15 expected SNP genotypes were detected (Table 2).
The combination of the five mutations in the MC1R gene resulted in eight different haplotypes deposited in GenBank under the accession numbers KU705368-KU705368 (Table 3). The dominant black Ed haplotype H3 (AATGT) was found only in Sawaknee and Najdi sheep (Table 4). The haplotypes H1 (TGCTC), H2 (TGTGT), and H6 (AGTGT) were represented in the three Saudi indigenous breeds. However, the four haplotypes, H4 (TGCGC), H5 (TGCTT), H7 (AGCTC), and H8 (TGTTT), were rare and represented in either one or two breeds (Table 4).
Based on the eight MC1R haplotypes recovered during this study, 36 expected haplotype combinations were constructed. Out of these expected combinations, 16 were observed in the studied sheep. Five out of the 16 MC1R observed haplotype combinations were homozygotes with a total number of 78 individuals. The remaining 11 combinations were represented by 30 animals. Generally, the haplotype combinations were not evenly distributed among the three Saudi breeds (Figure 2). The only haplotype combination that included individual sheep from the three Saudi breeds was the homozygous G8 (H2/H2) (Figure 2). The observed haplotype combinations (G3, G9, G11, and G12) carrying dominant Ed MC1R allele with the nonsynonymous mutations at 218 and 361 positions were mainly from Najdi and Sawaknee sheep (Figure 2). Amongst the 9 Najdi haplotype combinations, 6 were unique whereas 2 out of 7 Herri ones were unique (Figure 2). Out of the 6 Naeimi haplotype combinations, three were unique.
The MC1R protein is a highly polymorphic 7-transmembrane G protein-coupled receptor [16]. The MC1R protein is predominantly expressed in melanocytes and plays a central role in regulating melanin production in these specialized cells [8,17]. When the MC1R receptor is bound by its agonist, alpha melanocyte-stimulating hormone (α-MSH), the intracellular levels of cyclic adenosine monophosphate (cAMP) are elevated through a G-protein signaling pathway and the black/brown eumelanin is produced [18]. However, if the MC1R is bound by its antagonist, ASIP expressed by agouti gene, α-MSH binding is blocked, the cAMP levels are reduced and the red/yellow pheomelanin is produced [16,19]. Thus, by varying the levels of α-MSH and agouti protein, the wild type receptor can effectively switch melanin biosynthesis between its two forms [19–21].
In sheep, many loci contributing in coat color, e.g. agouti (A), extension (E), and irregular spotting (S), have been characterized [22,23]. Among these loci, A locus, encoding for ASIP protein, and E locus, encoding for MC1R receptor, are the most studied ones. Although the recessive allele (a) in the A locus can lead to black genotyped sheep, the dominant allele (Ed) in E locus is epistatic over A alleles and produces black genotypes [22,23]. There are other two alleles in the E locus, E+ (wild type) and e [5,12,24,25]. In 1999, Våge et al [9] identified two nonsynonymous mutations that cosegregated with the dominant Ed allele in sheep with black coat colors, the first at position 73 (Met73Lys) and the second at position 121 (Asp121Asn) of MC1R polypeptide. When these two mutations were introduced into mouse MC1R receptors, the Met73Lys mutation showed constitutive activation whereas the Asp121Asn mutation was required for high affinity ligand binding [9]. In this study, these two mutations were detected in Saudi Najdi, generally black, and Sudanese Sawaknee, generally brown, sheep. The Ed haplotype, AATGT, was recovered from either homozygous or heterozygous sheep. Generally, there was a complete association between the presence of Ed allele (AATGT) and black or brown coat color. This haplotype has been previously detected in many black sheep breed worldwide (Table 5). For example, AATGT haplotype was the most common in Chinese Minxian sheep breed expressing black-coat [15]. In addition, this haplotype was observed in Brazilian colored Creole and Italian black Massese sheep breeds [12,25]. Moreover, Ed allele has been observed in other colored sheep breeds including Aragonesa, Romney Marsh black-boned, Kazakh Fat-Rumped, and Xalda [10,15,26].
Although both Met73Lys and Asp121Asn mutations are required for dominant black sheep genotypes, haplotypes having only the Met73Lys mutation, H6 (AGTGT), and H7 (AGCTC), were detected in indigenous Saudi sheep expressing different coat colors. The haplotype H6 was more frequent than H7, in addition occurring in homozygous individuals. These haplotypes were not detected in other sheep breeds worldwide. Våge et al [9] showed that the Met73Lys mutation leads to a constitutive activation of MC1R in sheep. In this study, this mutation led to black coat sheep when it is observed in homozygous individuals, Najdi sheep with genotype H6/H6. However, its occurrence in heterozygous genotypes did not lead to complete black coats, e.g. white Herri with black nose and mouth. It has been reported that single mutations in MC1R gene can lead to either entirely or partially black animals [27–29]. For example, Kijas et al [27] identified a single nonsynonymous mutation at position 121 (Asp121Asn) of MC1R gene in pigs associated with both generally black colored and white or red pigs with black spots. Moreover, Kerje et al [28] showed that the Glu92Lys mutation at chicken MC1R is the causal mutation in black phenotypes of White Leghorn breed.
In addition to the 3 haplotypes of Ed allele, 5 of the wild type E+ allele were detected in Saudi sheep. Out of these 5 haplotypes, 3 (H4, H5, and H8) were rare among the Saudi sheep and only detected in Brazilian sheep [25]. However, the other two common haplotypes (H1 and H2) were detected in other sheep worldwide (Table 5). For example, H1 (TGCTC), the most common haplotype of E+ allele in Saudi sheep, was also the most common haplotype in Chinese Large-tailed Han white breed [15].
The present study supports the suggestion that the two independent nonsynonymous mutations (Met73Lys and Asp121Asn) in MC1R are associated with b pigmentation in sheep. Further studies are carried out to characterize the sequence variations in the lack or red coat colors in Saudi indi-genous Najdi and Sudanese sheep breeds, respectively. Moreover, the Met73Lys mutation was observed in Saudi sheep and it is suggested to be associated with partial black or red pigmentation in sheep. Further studies are carried out to characterize the sequence variations in the agouti locus and their correlation with MC1R variations will be assessed in Saudi sheep breeds.
ACKNOWLEDGMENTS
This project was supported by the Deanship of Scientific Research through the research group project RGP-028, King Saud University, Saudi Arabia.
Figure 1
Illustration of sheep coat colors in the studied four breeds. (A) Najdi sheep with Black coat; (B) Herri sheep with white coat; (C) Naeimi sheep with brownish white coat and brown face; (D) Sawaknee sheep with brown coat.

Figure 2
Distribution of different melanocortin-1 receptor (MC1R) haplotype combinations, G1 (H1/H1), G2 (H1/H2), G3 (H1/H3), G4 (H1/H4), G5 (H1/H5), G6 (H1/H6), G7 (H1/H7), G8 (H2/H2), G9 (H2/H3), G10 (H2/H6), G11 (H3/H3), G12 (H3/H6), G13 (H4/H4), G14 (H4/H5), G15 (H6/H6), G16 (H7/H8), across Saudi and non-Saudi sheep breeds. The breed designations are as follows: NJ (Najdi), NM (Naeimi), HE (Herri) and SW (Sawaknee).
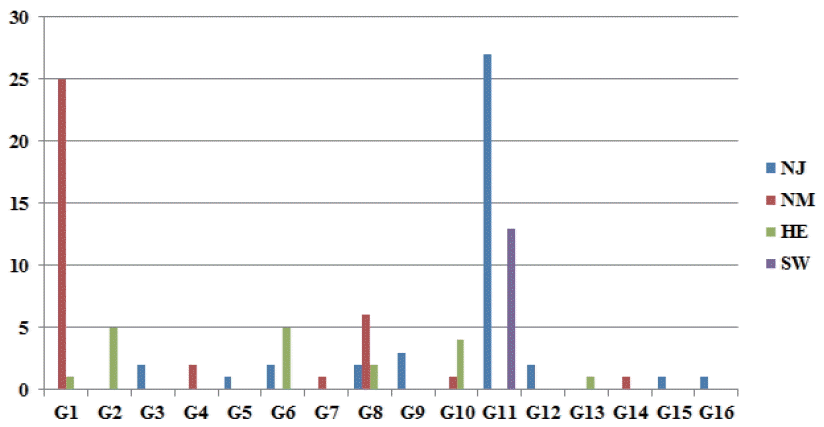
Table 1
Allele frequencies of the 5 SNPs of MC1R gene detected in Saudi and non-Saudi sheep breeds
Table 2
Constructed genotypes based on 5 SNPs of MC1R and their distribution in Saudi and non-Saudi sheep breeds
Table 3
The construction of different MC1R haplotypes recovered in this study and their corresponding E allele
Table 4
Distribution of MC1R haplotypes among Saudi and non-Saudi sheep breeds
| H1)/breed | H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | Total |
|---|---|---|---|---|---|---|---|---|---|
| Najdi | 5 | 7 | 61 | 0 | 1 | 6 | 1 | 1 | 82 |
| Naeimi | 53 | 13 | 0 | 3 | 1 | 1 | 1 | 0 | 72 |
| Herri | 12 | 13 | 0 | 2 | 0 | 9 | 0 | 0 | 36 |
| Sawaknee | 0 | 0 | 26 | 0 | 0 | 0 | 0 | 0 | 26 |
| Total | 70 | 33 | 87 | 5 | 2 | 16 | 2 | 1 | 216 |
Table 5
The 8 MC1R haplotypes recovered during this study and their distribution worldwide
REFERENCES
1. Adalsteinsson S. Colour inheritance in Icelandic sheep and relation between colour, fertility and fertilization. J Agric Res Iceland 1970; 2:3–135.
2. Abouheif MA, Abdo GM, Basmaeil SM, Alsobayel AA. Identification of the preference patterns of different breeds of sheep for consumption in Saudi Arabia. Asian-Australas J Anim Sci 1989; 2:129–32.

3. Andersson L. Melanocortin receptor variants with phenotypic effects in horse, pig, and chicken. Ann NY Acad Sci 2003; 994:313–8.


4. Chen SY, Huang Y, Zhu Q, et al. Sequence characterization of the MC1R gene in yak (Poephagus grunniens) breeds with different coat colors. J Biomed Biotechnol 2009; Article ID 861046


5. Searle AG. Comparative genetics of coat colour in mammals. NY, USA: Academic Press; 1968.
6. Bultman SJ, Michaud EJ, Woychik RP. Molecular characterization of the mouse agouti locus. Cell 1992; 71:1195–204.


7. Robbins LS, Nadeau JH, Johnson KR, et al. Pigmentation phenotypes of variant extension locus alleles result from point mutations that alter MSH receptor function. Cell 1993; 72:827–34.


8. Hunt G, Kyne S, Wakamatsu K, Ito S, Thody AJ. Nle4DPhe7 α-melanocyte-stimulating hormone increases the eumelanin: phaeomelanin ratio in cultured human melanocytes. J Investig Dermatol 1995; 104:83–5.


9. Våge DI, Klungland H, Lu D, Cone RD. Molecular and pharmacological characterization of dominant black coat color in sheep. Mamm Genome 1999; 10:39–43.


10. Våge DI, Fleet MR, Ponz R, et al. Mapping and characterization of the dominant black colour locus in sheep. Pigment Cell Res 2003; 16:693–7.


11. Fontanesi L, Beretti F, Riggio V, et al. Missense and nonsense mutations in melanocortin 1 receptor (MC1R) gene of different goat breeds: association with red and black coat colour phenotypes but with unexpected evidences. BMC Genet 2009; 10:47



12. Fontanesi L, Beretti F, Riggio V, et al. Sequence characterization of the melanocortin 1 receptor (MC1R) gene in sheep with different coat colours and identification of the putative e allele at the ovine Extension locus. Small Rumin Res 2010; 91:200–7.

13. Fang M, Larson G, Ribeiro HS, Li N, Andersson L. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet 2005:e1000341



14. Sambrook J, Fritsh EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edNY: Cold Spring Harbor Laboratory Press; 1989.
15. Yang GL, Fu DL, Lang X, et al. Mutations in MC1R gene determine black coat color phenotype in Chinese sheep. Sci World J 2013; Article ID 675382


16. Mountjoy KG, Robbins LS, Mortrud MT, Cone RD. The cloning of a family of genes that encode the melanocortin receptors. Science 1992; 257:1248–51.


17. Cone RD, Lu D, Koppula S, et al. The melanocortin receptors: agonists, antagonists, and the hormonal control of pigmentation. Recent Prog Horm Res 1996; 51:287–317.

18. Donatien PD, Hunt G, Pieron C, et al. The expression of functional MSH receptors on cultured human melanocytes. Arch Dermatol Res 1992; 284:424–26.


19. García-Borrón JC, Sánchez-Laorden BL, Jiménez-Cervantes C. Melanocortin-1 receptor structure and functional egulation. Pigment Cell Res 2005; 18:393–410.

20. Klungland H, Våge DI. Molecular genetics of pigmentation in domestic animals. Curr Genomics 2000; 1:223–42.

21. Klungland H, Våge DI. Pigmentary switches in domestic animal species. Ann NY Acad Sci 2003; 994:331–8.


22. Lauvergne JJ. Genetics of fleece coloring of three French sheep breeds: isolognot, bizet and berrichon. Ann Genet Sel Anim 1975; 7:263–76.


23. Lauvergne JJ, Hoogschagen P. Genetic formulas for the colour in the Texel, the Dutch and the Zwartbles sheep in the Netherlands. Ann Genet Sel Anim 1978; 10:343–51.



24. Sponenberg DP. Genetics of colour and hair texture. Piper L, Ruvinsky A, editorsThe genetics of sheep. New York: CAB International; 97:p. 51–86.

25. Hepp D, Gonçalves GL, Moreira GRP, et al. Identification of the e allele at the extension locus (MC1R) in Brazilian Creole sheep and its role in wool color variation. Genet Mol Res 2012; 11:2997–3006.


26. Royo LJ, Alvarez I, Arranz J, et al. Differences in the expression of the ASIP gene are involved in the recessive black coat colour pattern in sheep: evidence from the rare Xalda sheep breed. Anim Genet 2008; 39:290–3.


27. Kijas JMH, Wales R, Tornsten A, et al. Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics 1998; 150:1177–85.









 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





