 |
 |
| Anim Biosci > Volume 29(9); 2016 > Article |
|
Abstract
Adrenergic, alpha-1B-, receptor (ADRA1B) and peroxisome proliferator-activated receptor gamma, coactivator 1 beta (PPARGC1B) genes are involved in regulation of hen ovarian development. In this study, these two genes were investigated as possible molecular markers associated with hen-housed egg production, egg weight (EW) and body weight in Chinese Dagu hens. Samples were analyzed using the polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) technique, followed by sequencing analysis. Two novel single nucleotide polymorphisms (SNPs) were identified within the candidate genes. Among them, an A/G transition at base position 1915 in exon 2 of ADRA1B gene and a T/C mutation at base position 6146 in the 3′-untranslated region (UTR) of PPARGC1B gene were found to be polymorphic and named SNP A1915G and T6146C, respectively. The SNP A1915G (ADRA1B) leads to a non-synonymous substitution (aspartic acid 489-to-glycine). The 360 birds from the Dagu population were divided into genotypes AA and AG, allele A was found to be present at a higher frequency. Furthermore, the AG genotype correlated with significantly higher hen-housed egg production (HHEP) at 30, 43, 57, and 66 wks of age and with a higher EW at 30 and 43 wks (p<0.05). For the SNP T6146C (PPARGC1B), the hens were typed into TT and TC genotypes, with the T allele shown to be dominant. The TC genotype was also markedly correlated with higher HHEP at 57 and 66 wks of age and EW at 30 and 43 wks (p<0.05). Moreover, four haplotypes were reconstructed based on these two SNPs, with the AGTC haplotype found to be associated with the highest HHEP at 30 to 66 wks of age and with higher EW at 30 and 43 wks (p<0.05). Collectively, the two SNPs identified in this study might be used as potential genetic molecular markers favorable in the improvement of egg productivity in chicken breeding.
Egg productivity is the most important of economic traits in laying poultry (Kim et al., 2004). It includes egg production, egg weight (EW), sexual maturity and the number of recruited dominant follicles that are regulated by various endocrine, paracrine and autocrine factors (Su et al., 1996; Qin et al., 2015a). Many of the modern chicken breeds have been generated by conventional breeding methods (such as self-selection and family selection); with enhanced egg production traits a key focus (Qin et al., 2015b). However, egg productivity is inherited as a polygenic trait, with low to moderate heritability, making the level of genetic improvements difficult to estimate (Luo et al., 2007; Biscarini et al., 2010; Venturini et al., 2013). It has been proposed that a candidate gene approach is a cost-effective means of investigating associations of gene polymorphisms and quantitative trait loci responsible for variations in traits of interest (Rothschild and Soller, 1997; Linville et al., 2001). The identification and utilization of single nucleotide polymorphisms (SNP) in candidate genes associated with genotypes that have significant effects on economically important traits has become increasingly important in chicken breeding programs (Liu et al., 2010; Zhang et al., 2012; Qin et al., 2015b). This technique has attracted a growing number of animal breeding researchers in recent years. Nevertheless, more candidate genes and SNPs are required for marker-assisted selection in chicken breeding. Previous studies have demonstrated adrenergic, alpha-1B-, receptor (ADRA1B) and peroxisome proliferator-activated receptor gamma, coactivator 1 beta (PPARGC1B) genes are involved in the regulation of ovarian follicular development, selection and maturation in broilers and rats (Aguado and Ojeda, 1986; Tcherepanova et al., 2000; McDerment et al., 2012), with a potential effect on egg production in laying hens.
Gene ADRA1B encodes alpha-1B adrenergic receptor (α1B ADR) that is a member of the G-protein-coupled family of transmembrane receptors (Allen et al., 1991), located at chromosome 13 in chickens (Gene ID: 373890). Southern blot analysis and nucleotide sequencing indicate that this gene comprises two exons and a single large intron of at least 16 kb in rats or 20 kb in humans that interrupts the coding region at the end of the putative sixth transmembrane domain (Ramarao et al., 1992; Gao and Kunos, 1993). In chickens, due to alternative splicing of pre-mRNA following transcription of primary transcript mRNA, there are at least three spliced variants of ADRA1B transcript (Ping and Faber, 1993; Port and Bristow, 2001). Not only has the α1B ADR been shown to activate mitogenic response and regulate growth and proliferation of many cells (Waldrop et al., 2002; Kodama and Togari, 2013), but also it was suggested the receptor is implicated for follicle development, selection and maturation in hen ovaries (Aguado and Ojeda, 1986; McDerment et al., 2012).
The PPARGC1B gene encodes peroxisome proliferator-activated receptor gamma (PPARγ), coactivator 1 beta (PPARGC1B, or PGC-1β), it belongs to a small family of nuclear receptor coregulators that coordinate responses to metabolic stimuli and stressors (Handschin and Spiegelman, 2006; Lin, 2009). This gene is located at chromosome 13 and consists of 11 exons that encode 997 amino acids in chicken (Accession No.XP_414479), with its synonymous name as PPARγ coactivator 1 beta (PGC1B) gene, or PPARγ coactivator-1 (PGC-1) related estrogen receptor alpha coactivator (PERC) gene in human and mouse (Kressler et al., 2002; St-Pierre et al., 2003). It was shown that PGC-1, including PPARγ coactivator 1 alpha (PGC-1α) and PGC-1β, is a coactivator of estrogen receptor-alpha (ERα)-dependent transcriptional activity, which may serve as a convergence point between PPARγ and ERα signaling (Tcherepanova et al., 2000). The role of estrogens and ovarian estrogen receptors (ERα, β) in hen folliculogenesis, follicle maturation, ovulation and reproduction has been well established (Drummond et al., 1999; 2012; Hrabia et al., 2004; 2008; Brunström et al., 2009). Therefore, it was inferred that variations of PPARGC1B and ADRA1B gene sequences may greatly influence chicken ovarian development and egg laying performance, although the molecular mechanisms of both of them in regulating follicle development and growth remain unknown. While the Chinese Dagu chicken is an important animal resource, little characterization of ADRA1B and PPARGC1B genetic polymorphisms and possible correlations with egg performance in indigenous Chinese breeds has been performed (Park et al., 2006; Akhmetov et al., 2009; Lee et al., 2011).
In this study, ADRA1B and PPARGC1B fragments were examined for novel sequence polymorphism using a polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) approach and sequencing analysis. Associations between the newly identified genotypes and the egg production traits were explored in local Chinese Dagu hens. The aim of this work is to provide a potential genetic molecular marker able to facilitate selection for improved egg production traits in chicken breeding.
The Chinese Dagu chickens were provided by College of Animal Science and Technology of Jilin Agricultural University. As reported by Qin et al. (2015b), the eggs used for hatching were randomly selected from the Dagu chicken population, with 360 hens hatched and raised in layered batteries under the same rearing conditions, to include free access to water and feed in accordance to the nutrient requirements of local Chinese Dagu hens (NY/T 33-2004, Beijing, China). Approaching 16 wks of age, hens were reared in individual cages under constantly maintained conditions. All of the birds were exposed to a 16 L:8 D photoperiod, with lights on at 5:00 am. After the start of laying, eggs were collected and recorded daily, with EWs determined on one day in a week. Body weight was recorded following feed and water restrictions at 30 and 43 wks of age, with the individual laying performance calculated. Egg production traits examined in this study included hen-housed egg production (egg laying number) at 30, 43, 57, and 66 wks of age, EW, and body weight at 30 and 43 wks of age. All animal experiments were performed in accordance with laws of the People’s Republic of China regarding animal protection.
A total of 360 individuals from Chinese Dagu chickens were sampled in this work. For each bird, about 1 mL of blood was collected from the wing vein at 300 days of age and DNA was extracted using a standard phenol-chloroform method. To detect genomic DNA purity, 1% agarose gel electrophoresis and ultraviolet-spectrophotometer assay were performed, with final concentrations between 2 and 10 ng/μL detected. Primers were designed based the ADRA1B (GenBank accession No. XM_414483.3) and PPARGC1B (Accession No. XM_414479.4) gene sequences in chicken. The primer pairs used to amplify the fragments for ADRA1B (forward 5′-TTAACAGAGCGCAACGGACA-3′; reverse 5′-ACAATAGCTGAATAATTGGCTT-3′) and PPARGC1B gene (forward 5′-GTAGGAAAGGAGCC GATGT-3′; reverse 5′-AACGCCAGAAGGAAAGAAT-3′) were screened after examination in advance.
PCR reactions were performed in a total volume of 50 μL, including: 20 μL of 2X Taq Master Mix, 100 nM of each primer, 25 to 50 ng of template DNA, and 22 μL RNase-free Water. The PCR conditions included: 94°C for 2 min, followed by 35 cycles at 94°C for 30 s for denaturing, 59°C (55°C) for 30 s for annealing of ADRA1B gene (PPARGC1B gene), 72°C for 30 s for extension, and a final extension at 72°C for 2 min.
PCR products were purified with the Wizard prep PCR purification system (Promega, Madison, WI, USA), cloned into the Promega pGEM-T easy vector according to the methods published by Sambrook and Russell (2001), and the obtained fragments commercially sequenced. Two independent PCR amplifications were performed for each sampled bird, with sequences analyzed using BLAST (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html) to confirm the expected chicken ADRA1B and PPARGC1B gene sequences. The obtained sequences were then aligned using the DNAMAN software version 6.0 to identify nucleotide substitutions.
To screen for ADRA1B and PPARGC1B gene polymorphism, the confirmed PCR products were further analyzed using the SSCP assay as previously described (Qin et al., 2015b). Briefly, Every 10 μL PCR product was mixed with 6 μL of loading dye (0.025% bromophenol blue, 0.025% xylene cyanol, 98% deionized formamide, 2% glycerin and 10 mM ethylenediaminetetraacetic acid [EDTA]). Denatured at 99°C for 10 min beforehand, the samples were cooled rapidly on ice, and then loaded on 10% polymerized gels (acrylamide:bisacrylamide, 39:1) of size 16 cm×18 cm. Electrophoresis was carried out at 130 V for 7 to 10 h at room temperature in 1×tris-borate-EDTA (TBE) buffer. After silver stain, the gels were examined under upper white by gel photography system (GeneSnap from SynGene). To avoid false positive/negative results due to artificial manipulation in the experiment, each sample was confirmed by repeated amplifications and detections. Haplotypes were reconstructed according to the genotyping data obtained from all 360 individuals with the PHASE program (Stephens et al., 2001).
Genotype and allelic frequencies at each SNP site were calculated, with each polymorphism evaluated for Hardy-Weinberg equilibrium using a Pearson’s goodness-of-fit chi-square test (degree of freedom = 1). Gene homozygosity (Ho), heterozygosity (He), effective number of alleles (Ne) and polymorphism information content (PIC) were statistically analyzed using the POPGENE v. 1.32 software (Yeh et al., 1997).
Associations of single polymorphisms or haplotypes with laying performance traits were analyzed using general linear model procedure in SPSS 18.0. The model was as below:
where Yijk is the phenotypic value of the target trait, such as egg laying number, μ is the population mean, Li is the fixed effect of the line, Gj is the fixed effect of the SNP genotype or haplotype, Fk is the random effect of the family and eijk are the residuals. Type III sum of squares was used in each test. Values were considered significant at p<0.05 and presented as least square means±standard errors.
A 389 bp PCR amplicon obtained for ADRA1B gene and a 276 bp fragment for PPARGC1B were cloned using specifically designed primers, with the obtained fragments sequenced. Comparisons of the predicted sequences with the corresponding GenBank sequences (XM_414483.3 for ADRA1B and XM_414479.4 for PPARGC1B gene) were performed using the BLAST software provided by the NCBI server. Primer specificity was confirmed, with PCR amplicon sizes corresponding to the expected sequences of the candidate genes (Figure 1), and further confirmed by comparing the cloned PCR product sequences with the direct genomic PCR products from the same individual. For all of the examined birds, no more than two allelic sequences were observed, suggesting that the primer pair specifically amplified a single gene.
In the study, genotyping of the amplified ADRA1B and PPARGC1B target fragments was conducted by PCR-SSCP analysis. The typing results showed that there were two genotypes (AA and AG) in the coding region of ADRA1B gene (Figure 2) and two genotypes (TT and TC) in the 3′-untranslated region (UTR) of PPARGC1B gene within the Dagu hen population (Figure 3). Haplotype reconstruction was performed based on these genotype data and four haplotypes (AATT, AATC, AGTT, and AGTC) were identified among the 360 individual hens examined. The haplotype present at the highest frequency was the AATT haplotype (0.55), with the AGTT haplotype the next most frequent (0.18), followed by AATC (0.17), and AGTC (0.10).
Based on the PCR-SSCP banding patterns of the ADRA1B and PPARGC1B fragments, polymorphism of the target sequences was revealed following sequencing and aligning of the PCR products. For the ADRA1B fragment, an A/G transition at base position 1915 in exon 2 of the coding region was detected (Figure 2), and named SNP A1915G. Furthermore, this noted A/G transition leads to a non-synonymous substitution, aspartic acid 489-to-glycine. For this SNP, the birds sampled were typed as either AA or AG genotypes by PCR-SSCP analysis. In PPARGC1B gene, a T/C transition at base position 6146 in the 3′-UTR was identified and named SNP T6146C (Figure 3), with birds typed as either TT or TC following PCR-SSCP analysis.
Genotypic and allelic frequencies for the ADRA1B and PPARGC1B genes are given in Table 1. For the SNP A1915G of ADRA1B gene, the frequency of allele A was notably higher than allele G, with the frequency of genotype AA higher than genotype AG in this population. For the SNP T6146C of PPARGC1B gene, the frequency of allele T was higher than allele C, with the frequency of genotype TT higher than genotype TC. The polymorphism at both sites, A1915G of ADRA1B and T6146C of PPARGC1B, were evaluated and found to exhibit significant genetic disequilibrium between the A and G alleles of ADRA1B and the T and C alleles of PPARGC1B (p<0.05).
As shown in Table 2, gene homozygosity (Ho) was higher than gene heterozygosity (He) for both the SNP A1915G (P1) of ADRA1B or for the SNP T6146C (P2) of PPARGC1B, with effective allele numbers of 1.314 (P1) and 1.288 (P2). The value of PIC for He in P1 (ADRA1B gene) was higher than that of P2 (PPARGC1B gene), but the polymorphism was not higher and only varied from moderate (0.210) to low (0.198).
The SNP A1915G (ADRA1B) genotype AG was significantly associated with higher hen-housed egg production (HHEP) at 30, 43, 57, and 66 wks of age and with EW at 30 and 43 wks (Table 3; p<0.05). For the SNP T6146C (PPARGC1B), genotype TC was markedly correlated with higher HHEP at 57 and 66 wks of age and with EW at 30 and 43 wks (Table 3; p<0.05). However, no significant difference was observed between AA and AG or between TT and TC regarding body weight at 30 and 43 wks (p>0.05).
Among the four haplotypes, haplotype AGTC was found to be correlated with the highest HHEP at 30 to 66 wks of age and highest EW at 30 and 43 wks (Table 4; p<0.05), followed by haplotype AGTT and AATC associated with the higher HHEP at 43 to 66 wks of age, while with highest EW at 43 wks (p<0.05); and with the lowest levels associated with haplotype AATT (p<0.05). Concerning body weight at 30 and 43 wks, there were no significant differences between the four haplotypes (p>0.05).
China has a wide variety of indigenous chicken resources. Innovatively improving and utilizing the egg production trait is becoming one of the important tasks in the Chinese local chicken industry. However, egg production is inherited as a polygenic trait, with low to moderate heritability (Luo et al., 2007; Biscarini et al., 2010; Venturini et al., 2013), making genetic improvement of this trait more costly based on estimated breeding values, but poorly effective by using traditional breeding method (Qin et al., 2015b). Therefore, genetic factors are destined to play a pivotal role in promoting egg production traits to further this economically important resource. Involved in the process of egg production are not only members of the glycoprotein hormone family of gonadotropins, such as follicle-stimulating hormone (FSH) and luteinizing hormone (LH), but also a wide variety of cellular and tissue-level signal transductions that play critical roles in regulating normal follicular development and oocyte maturation (Qin et al., 2015a). These processes are also controlled by many local intraovarian factors in an autocrine or/and paracrine manner, such as the α1B ADR and PGC-1β proteins (Aguado and Ojeda, 1986; Tcherepanova et al., 2000; McDerment et al., 2012). Polymorphisms associated with egg production related hormones, growth factors, and sex hormones such as FSH beta subunit (FSHb), LH, prolactin (PRL), growth hormone (GH), transcription factor forkhead box L2 (FOXL2) and growth differentiation factor-9 (GDF9) have been intensively studied in chickens (Feng et al., 1997; Cui et al., 2006; Onagbesan et al., 2006; Qin et al., 2015b). However, little is known regarding polymorphisms in chicken ADRA1B and PPARGC1B genes (Park et al., 2006; Akhmetov et al., 2009; Lee et al., 2011). In an attempt to identify novel DNA markers associated with egg production traits in chickens, we examined polymorphisms in ADRA1B and PPARGC1B and evaluated their associations with egg production traits in Chinese local hens.
The present work, was the first to provide information about genetic polymorphisms in ADRA1B and PPARGC1B genes in chickens. The A/G transition in exon 2 of ADRA1B gene was identified to bring about a non-synonymous substitution, aspartic acid 489-to-glycine, in the deduced amino acid sequence of ADRA1B receptor. The substitution of amino acid residue was located in intracellular region of the carboxy terminus tail of the receptor (Piirainen et al., 2011). Previous studies in mammals and humans have elucidated the structural feature of the region that is implicated to the intracellular signal transduction of this receptor (Eason and Liggett, 1995; Jewell-Motz et al., 1997; Olah and Stiles, 1998; Shim et al., 2002). It is possible that sequence substitution may influence overall receptor architecture and subsequently disrupt the conformation of additional domains (Jewell-Motz et al., 1997; Olah and Stiles, 2000; Scheer et al., 2000). However, the possible functional alterations of ADRA1B receptor caused by the A/G mutation was unclear, and the potential varied effects on the ovarian follicle development and egg production by the change remain to be further explained in chicken. As for the transition of a T/C at base position 6146 in the 3′-UTR of PPARGC1B gene, it is reasonable to assume that a possible MicroRNA (miRNA) regulatory change might occur with the presence of this mutation in the 3′-UTR (Hossain et al., 2009; Yao et al., 2009; Yang et al., 2013), as any mutations present in the binding sites may reverse the biological effect of the target gene on hen ovary development and growth. In recent years, several miRNAs localized in the first intron of the PPARGC1B gene and in the regulatory region flanking the gene’s transcription start site in mouse and rat, such as miR-182/96/183, miR-205, miR-378 and miR-378*, were identified to attenuate the function of PGC-1β protein that is a coactivator of the ERα signaling (Tcherepanova et al., 2000; John et al., 2012; Mallat et al., 2014). Nevertheless, the investigation for a candidate regulator of endogenous miRNA targeting the site in the 3′-UTR of PPARGC1B gene is ongoing.
In this study, the two novel SNPs in the hen ADRA1B and PPARGC1B fragments were found to significantly correlate with the heightened HHEP and the increased EW (p<0.05). While these two candidate genes have been previously implicated in follicular development (Aguado and Ojeda, 1986; Tcherepanova et al., 2000; Drummond and Fuller, 2012); McDerment et al., 2012), and it indicated that the two candidates may be involved in the regulation of chicken egg productivity. Furthermore, association analyses on the four haplotypes (AATT, AGTT, AATC, and AGTC) have shown that the ADRA1B and PPARGC1B polymorphisms are significantly associated with egg production traits in Dagu chickens. Therefore, these results indicate that the two SNPs identified in ADRA1B and PPARGC1B might serve as possible molecular markers to aid in the improvement of egg production traits in chicken breeding.
In addition, we demonstrated that the frequency of allele A is predominantly higher than allele G for the SNP A1915G (ADRA1B) and the frequency of allele T was significantly higher than that of allele C for the SNP T6146C (PPARGC1B). Moreover, the gene homozygosity (Ho) was higher than gene heterozygosity (He) for the two SNPs. Both of these SNPs were found to be under genetic disequilibrium possibly due to allele A (ADRA1B) or allele T (PPARGC1B) being naturally one of the predominant alleles during genetic evolution, thus being more conserved and more common than other alleles in this population. However, the allele G or allele C with lower allelic frequency was associated with the heightened HHEP and the EW (p<0.05). The reason why this phenomenon occurred may mainly be explained by the following two aspects: i) the mutation A1915G in ADRA1B or T6146C in PPARGC1B was initially present in the original chicken population at a lower frequency; ii) they are novel mutants. Even if the allele (G or C) was correlated with improved egg production traits, a natural or/and artificial selection pressure was not enough to increase the frequency to a higher level within a very limited number of generations. Certainly, the egg production traits of local Dagu populations have been selected with the aim of enhancing early sexual maturity, egg laying numbers and EW traits for the last six generations. Additionally, it cannot be ignored that the number of birds examined in each population was not enough to demonstrate the true frequency and an extreme allele frequency was estimated as a result. Then, if the reason is correct, the genetic disequilibrium was easy to be understood as aforementioned. As for the SNP T6146C occurring in the 3′-UTR of PPARGC1B gene, the effect might be due to a tight linkage with either an advantageous allele or with an artificially selected economically favorable trait, such as higher egg production. Collectively, the results of the present study strongly suggest that the two novel ADRA1B and PPARGC1B polymorphisms are associated with egg production and EW, thus are potential molecular markers for egg productivity in Chinese Dagu chicken breeding.
ACKNOWLEDGMENTS
The work was supported by the National Natural Science Funds (No. 31272431), the National High Technology Research and Development Program of China (No. 2011AA100305), the China Agriculture Research System (No. CARS-42), the Jilin Provincial Agriculture Research System, and the Key Project of Education Department of Jilin Province (No. 2013-47).
Figure 1
Amplification of the fragments of chicken ADRA1B and PPARGC1B genes. M, DL 2000 DNA marker; Lanes 1 to 3, fragments of ADRA1B gene amplified; Lanes 4 to 6, fragments of PPARGC1B gene amplified. ADRA1B: adrenergic, alpha-1B-, receptor; PPARGC1B: peroxisome proliferator-activated receptor gamma, coactivator 1 beta.

Figure 2
PCR-SSCP band patterns at the A1915G site of the chicken ADRA1B gene fragment. (a) The capital letters (AA and AG) on the top indicate the different genotypes at the SNP locus by PCR-SSCP. (b) The A/G transition at base position 1915 in the exon 2 of ADRA1B gene (Accession No. XM_414483.3) was found by sequencing and alignment. The bird with homotype A1915A was classified as genotype AA, and the heterotype of A1915G named genotype AG basing on the band patterns of the PCR-SSCP and the sequencing information. PCR-SSCP: polymerase chain reaction-single strand conformation polymorphism; ADRA1B: adrenergic, alpha-1B-, receptor; SNP, single nucleotide polymorphism.
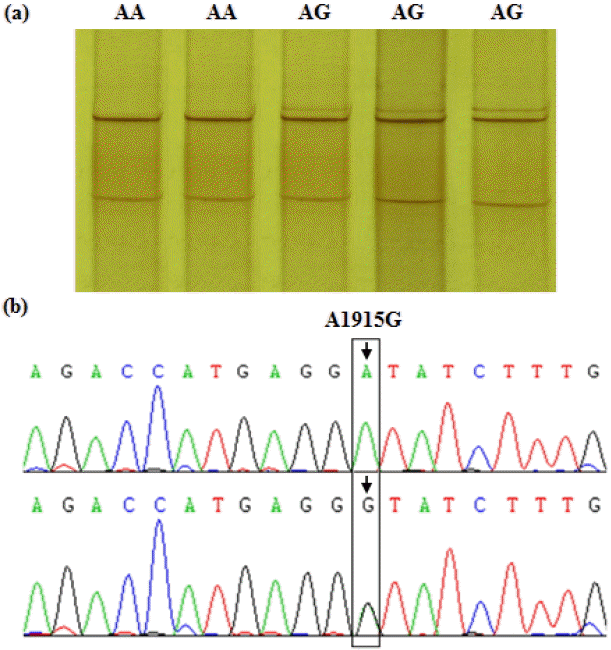
Figure 3
PCR-SSCP band patterns at the T6146C site of the chicken PPARGC1B gene fragment. (a) The capital letters (TT and TC) on the top indicate the different genotypes at the SNP locus by PCR-SSCP. (b) The T/C transition at base position 6146 in the 3′-UTR of PPARGC1B gene (Accession No. XM_414479.4) was detected by sequencing and alignment. The bird with homotype T6146T was termed genotype TT, and the heterotype of T6146C named genotype TC. PCR-SSCP: polymerase chain reaction-single strand conformation polymorphism; PPARGC1B: peroxisome proliferator-activated receptor gamma, coactivator 1 beta; SNP, single nucleotide polymorphism.
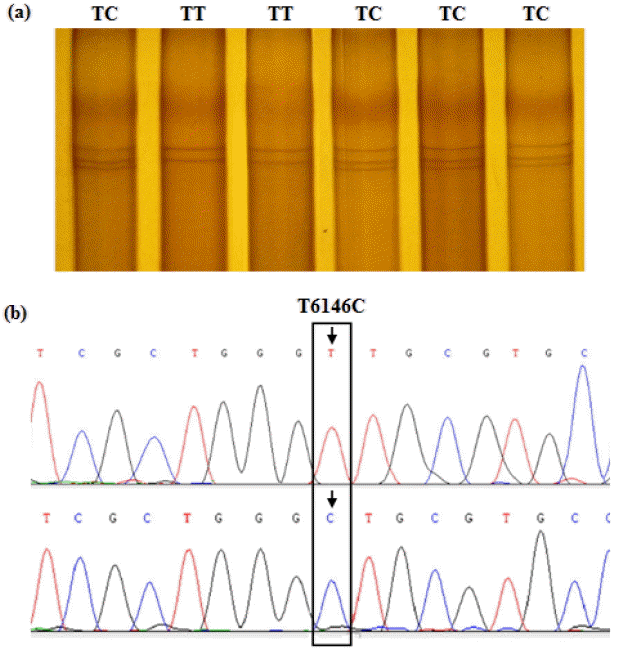
Table 1
Genotypic and allelic frequency at the SNP locus of ADRA1B and PPARGC1B genes in the Dagu chicken population
| SNP | Genotype | No. of chickens | Genotype frequency | Allele | Allele frequency | x2 |
|---|---|---|---|---|---|---|
| A1915G (ADRA1B) | AA | 260 | 0.722 | A | 0.861 | 12.69 * |
| AG | 100 | 0.278 | G | 0.139 | ||
| T6146C (PPARGC1B) | TT | 268 | 0.744 | T | 0.872 | 7.70* |
| TC | 92 | 0.256 | C | 0.128 |
Table 2
Polymorphism information analysis of chicken ADRA1B and PPARGC1B genes in the Chinese local Dagu chicken population
Table 3
Association of the polymorphism in chicken ADRA1B and PPARGC1B genes with egg production trait in the local Dagu hen population
| Egg production trait | Genotypes | |||
|---|---|---|---|---|
|
|
||||
| ADRA1B | PPARGC1B | |||
|
|
|
|||
| AA (260) (X̄ ± SE) | AG (100) (X̄ ±SE) | TT (268) (X̄ ±SE) | TC (92) (X̄ ±SE) | |
| Body weight at 30 wks (kg) | 2.79±0.43 | 2.77±0.55 | 2.80±0.04 | 2.73±0.05 |
| Body weight at 43 wks (kg) | 2.94±0.70 | 3.04±0.10 | 3.01±0.06 | 2.78±0.18 |
| Hen-housed egg production at 30 wks (No.) | 19.00±2.30a | 24.69±2.10b | 22.43±2.05 | 23.89±2.75 |
| Hen-housed egg production at 43 wks (No.) | 88.97±4.41a | 96.09±5.81b | 89.19±4.22 | 93.48±5.93 |
| Hen-housed egg production at 57 wks (No.) | 125.73±5.40a | 137.74±5.82b | 126.43±5.19a | 136.62±5.54b |
| Hen-housed egg production at 66 wks (No.) | 137.45±7.48a | 151.73±6.49b | 137.41±7.06a | 153.52±7.19b |
| Egg weight at 30 wks (g) | 55.25±0.54a | 56.97±0.59b | 55.02±0.57a | 56.89±0.55b |
| Egg weight at 43 wks (g) | 57.45±0.56a | 60.17±0.64b | 57.38±0.61a | 59.84±0.59b |
ADRA1B: adrenergic, alpha-1B-, receptor; PPARGC1B: peroxisome proliferator-activated receptor gamma, coactivator 1 beta; SE, standard error.
Table 4
Association of different haplotype polymorphism in chicken ADRA1B and PPARGC1B genes with egg production trait in the local Dagu hen population
| Egg production trait | Haplotype | |||
|---|---|---|---|---|
|
|
||||
| AATT (198) (X̄ ±SE) | AATC (61) (X̄ ±SE) | AGTT (66) (X̄ ±SE) | AGTC (35) (X̄ ±SE) | |
| Body weight at 30 wks (kg) | 2.79±0.06 | 2.70±0.06 | 2.82±0.07 | 2.81±0.09 |
| Body weight at 43 wks (kg) | 3.01±0.08 | 2.92±0.13 | 3.02±0.09 | 2.98±0.24 |
| Hen-housed egg production at 30 wks (No.) | 20.64±2.50a | 22.00±2.48a | 19.93±3.27a | 25.00±3.35 b |
| Hen-housed egg production at 43 wks (No.) | 86.62±4.95a | 94.50±6.96b | 93.40±5.7b | 102.13±7.25c |
| Hen-housed egg production at 57 wks (No.) | 123.27±7.28a | 130.86±8.59b | 133.93±8.59b | 145.50±8.78c |
| Hen-housed egg production at 66 wks (No.) | 133.18±8.49a | 147.64±9.88b | 148.80±8.44b | 160.63±9.74c |
| Egg weight at 30 wks (g) | 55.87±0.62a | 56.27±0.89ab | 57.57±1.10ab | 58.55±0.97b |
| Egg weight at 43 wks (g) | 57.94±0.64a | 59.92±0.73b | 60.89±0.97b | 60.39±1.06b |
REFERENCES
Aguado L, Ojeda SR. 1986. Prepubertal rat ovary: Hormonal modulation of beta-adrenergic receptors and of progesterone response to adrenergic stimulation. Biol Reprod 34:45–50.


Akhmetov II, Popov DV, Missina SS, Vinogradova OL, Rogozkin VA. 2009. The analysis of PPARGC1B gene polymorphism in athletes. Ross Fiziol Zh Im I M Sechenova 95:1247–1253.

Allen LF, Lefkowitz RJ, Caron MG, Cotecchia S. 1991. G-protein-coupled receptor genes as protooncogenes: Constitutively activating mutation of the alpha 1B-adrenergic receptor enhances mitogenesis and tumorigenicity. Proc Natl Acad Sci USA 88:11354–11358.



Biscarini F, Bovenhuis H, Ellen ED, Addo S, van Arendonk JAM. 2010. Estimation of heritability and breeding values for early egg production in laying hens from pooled data. Poult Sci 89:1842–1849.


Brunström B, Axelsson J, Mattsson A, Halldin K. 2009. Effects of estrogens on sex differentiation in Japanese quail and chicken. Gen Comp Endocrinol 163:97–103.


Cui JX, Du HL, Liang Y, Deng XM, Li N, Zhang XQ. 2006. Association of polymorphisms in the promoter region of chicken prolactin with egg production. Poult Sci 85:26–31.


Drummond AE, Baillie AJ, Findlay JK. 1999. Ovarian estrogen receptor alpha and beta mRNA expression: Impact of development and estrogen. Mol Cell Endocrinol 149:153–161.


Drummond AE, Fuller PJ. 2012. Ovarian actions of estrogen receptor-β: An update. Semin Reprod Med 30:32–38.


Eason MG, Liggett SB. 1995. Identification of a Gs coupling domain in the amino terminus of the third intracellular loop of the alpha 2A-adrenergic receptor. Evidence for distinct structural determinants that confer Gs versus Gi coupling. J Biol Chem 270:24753–24760.


Feng XP, Kuhnlein U, Aggrey SE, Gavora JS, Zadworny D. 1997. Trait association of genetic markers in the growth hormone and the growth hormone receptor gene in a White Leghorn strain. Poult Sci 76:1770–1775.


Gao B, Kunos G. 1993. Isolation and characterization of the gene encoding the rat alpha 1B adrenergic receptor. Gene 131:243–237.


Handschin C, Spiegelman BM. 2006. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev 27:728–735.


Hossain MM, Ghanem N, Hoelker M, Rings F, Phatsara C, Tholen E, Schellander K, Tesfaye D. 2009. Identification and characterization of miRNAs expressed in the bovine ovary. BMC Genomics 10:443



Hrabia A, Ha Y, Shimada K. 2004. Expression of estrogen receptor alpha mRNA in theca and granulosa layers of the ovary in relation to follicular growth in quail. Folia Biol (Krakow) 52:191–195.


Hrabia A, Wilk M, Rzasa J. 2008. Expression of alpha and beta estrogen receptors in the chicken ovary. Folia Biol (Krakow) 56:187–191.


Jewell-Motz EA, Donnelly ET, Eason MG, Liggett SB. 1997. Role of the amino terminus of the third intracellular loop in agonist-promoted down-regulation of the alpha-2A adrenergic receptor. Biochemistry 36:8858–8863.


John E, Wienecke-Baldacchino A, Liivrand M, Heinäniemi M, Carlberg C, Sinkkonen L. 2012. Dataset integration identifies transcriptional regulation of microRNA genes by PPARγ in differentiating mouse 3T3-L1 adipocytes. Nucl Acids Res 40:4446–4460.



Kim MH, Seo DS, Ko Y. 2004. Relationship between egg productivity and insulin-like growth factor-I genotypes in Korean native Ogol chickens. Poult Sci 83:1203–1208.


Kodama D, Togari A. 2013. Noradrenaline stimulates cell proliferation by suppressing potassium channels via G(i/o) -protein-coupled α(1B) -adrenoceptors in human osteoblasts. Br J Pharmacol 168:1230–1239.



Kressler D, Schreiber SN, Knutti D, Kralli A. 2002. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J Biol Chem 277:13918–13925.


Lee SH, Jang AS, Woo Park S, Park JS, Kim YK, Uh ST, Kim YH, Chung IY, Park BL, Shin HD, Park CS. 2011. Genetic effect of single-nucleotide polymorphisms in the PPARGC1B gene on airway hyperreactivity in asthmatic patients. Clin Exp Allergy 41:1533–1544.


Lin JD. 2009. Minireview: the PGC-1 coactivator networks: chromatin-remodeling and mitochondrial energy metabolism. Mol Endocrinol 23:2–10.



Linville RC, Pomp D, Johnson RK, Rothschild MF. 2001. Candidate gene analysis for loci affecting litter size and ovulation rate in swine. J Anim Sci 79:60–67.


Liu WJ, Sun DX, Yu Y, Li G, Tang SQ, Zhang Y, Wang YC, Zhang Y. 2010. Association of Janus kinase 2 polymorphisms with growth and reproduction traits in chickens. Poult Sci 89:2573–2579.


Luo PT, Yang RQ, Yang N. 2007. Estimation of genetic parameters for cumulative egg numbers in a broiler dam line by using a random regression model. Poult Sci 86:30–36.


Mallat Y, Tritsch E, Ladouce R, Winter DL, Friquet B, Li Z, Mericskay M. 2014. Proteome modulation in H9c2 cardiac cells by microRNAs miR-378 and miR-378. Mol Cell Proteomics 13:18–29.



McDerment NA, Wilson PW, Waddington D, Dunn IC, Hocking PM. 2012. Identification of novel candidate genes for follicle selection in the broiler breeder ovary. BMC Genomics 13:494



Olah ME, Stiles GL. 1998. Adenosine receptor-mediated signal transduction. Effects of Extracellular Adenosine and ATP on Cardiac Myocytes. Pelleg A, Belardinelli L, editorsR. G. Landes Company; Austin, TX, USA.: p. 7–38.
Olah ME, Stiles GL. 2000. The role of receptor structure in determining adenosine receptor activity. Pharmacol Ther 85:55–75.


Onagbesan OM, Metayer S, Tona K, Williams J, Decuypere E, Bruggeman V. 2006. Effects of genotype and feed allowance on plasma luteinizing hormones, follicle-stimulating hormones, progesterone, estradiol levels, follicle differentiation, and egg production rates of broiler breeder hens. Poult Sci 85:1245–1258.


Park KS, Shin HD, Park BL, Cheong HS, Cho YM, Lee HK, Lee JY, Lee JK, Kim HT, Park CS, Han BG, Kimm K, Oh B. 2006. Putative association of peroxisome proliferator-activated receptor gamma co-activator 1beta (PPARGC1B) polymorphism with Type 2 diabetes mellitus. Diabet Med 23:635–642.


Piirainen H, Ashok Y, Nanekar RT, Jaakola VP. 2011. Structural features of adenosine receptors: from crystal to function. Biochim Biophys Acta 1808. 1233–1244.


Ping P, Faber JE. 1993. Characterization of alpha-adrenoceptor gene expression in arterial and venous smooth muscle. Am J Physiol 265:H1501–H1509.


Port JD, Bristow MR. 2001. Altered beta-adrenergic receptor gene regulation and signaling in chronic heart failure. J Mol Cell Cardiol 33:887–905.


Qin N, Fan XC, Zhang YY, Xu XX, Tyasi TL, Jing Y, Mu F, Wei ML, Xu RF. 2015a. New insights into implication of the SLIT/ROBO pathway in the prehierarchical follicle development of hen ovary. Poult Sci 94:2235–2246.


Qin N, Liu Q, Zhang YY, Fan XC, Xu XX, Lv ZC, Wei ML, Jing Y, Mu F, Xu RF. 2015b. Association of novel polymorphisms of forkhead box L2 and growth differentiation factor-9 genes with egg production traits in local chinese Dagu hens. Poult Sci 94:88–95.


Ramarao CS, Denker JM, Perez DM, Gaivin RJ, Riek RP, Graham RM. 1992. Genomic organization and expression of the human alpha 1B-adrenergic receptor. J Biol Chem 267:21936–21945.


Rothschild MF, Soller M. 1997. Candidate gene analysis to detect genes controlling traits of economic importance in domestic livestock. Probe 8:13–20.
Sambrook J, Russell DW. 2001. Molecular Cloning: A Laboratory Manual. 3rd ednCold Spring Harbor Laboratory Press; New York, USA:
Scheer A, Costa T, Fanelli F, De Benedetti PG, Mhaouty-Kodja S, Abuin L, Nenniger-Tosato M, Cotecchia S. 2000. Mutational analysis of the highly conserved arginine within the Glu/Asp-Arg-Tyr motif of the alpha(1b)-adrenergic receptor: effects on receptor isomerization and activation. Mol Pharmacol 57:219–231.

Shim JO, Shin CY, Lee TS, Yang SJ, An JY, Song HJ, Kim TH, Huh IH, Soh UD. 2002. Signal transduction mechanism via adenosine A1 receptor in the cat esophageal smooth muscle cells. Cell Signal 14:365–372.


Stephens M, Smith NJ, Donnelly P. 2001. A new statisticalmethod for haplotype reconstruction frompopulation data. Am J Hum Genet 68:978–989.



St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. 2003. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem 278:26597–26603.


Su H, Silversides FG, Villeneuve P. 1996. Effects of sex-linked imperfect albinism (sal-s) in the chicken on the relationships of plasma concentrations of progesterone and17 beta-estradiol with egg production. Poult Sci 75:13–19.


Tcherepanova I, Puigserver P, Norris JD, Spiegelman BM, McDonnell DP. 2000. Modulation of estrogen receptor-alpha transcriptional activity by the coactivator PGC-1. J Biol Chem 275:16302–16308.


Venturini GC, Savegnago RP, Nunes BN, Ledur MC, Schmidt GS, El Faro L, Munari DP. 2013. Genetic parameters and principal component analysis for egg production from White Leghorn hens. Poult Sci 92:2283–2289.


Waldrop BA, Mastalerz D, Piascik MT, Post GR. 2002. alpha(1B)- and alpha(1D)-Adrenergic receptors exhibit different requirements for agonist and mitogen-activated protein kinase activation to regulate growth responses in rat 1 fibroblasts. J Pharmacol Exp Ther 300:83–90.


Yang S, Wang S, Luo A, Ding T, Lai Z, Shen W, Ma X, Cao C, Shi L, Jiang J, Rong F, Ma L, Tian Y, Du X, Lu Y, Li Y, Wang S. 2013. Expression patterns and regulatory functions of microRNAs during the initiation of primordial follicle development in the neonatal mouse ovary. Biol Reprod 89:126


Yao N, Lu CL, Zhao JJ, Xia HF, Sun DG, Shi XQ, Wang C, Li D, Cui Y, Ma X. 2009. A network of miRNAs expressed in the ovary are regulated by FSH. Front Biosci (Landmark Ed) 14:3239–3245.


Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX. 1997. POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta; Edmonton, Alberta, Canada:





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





